Aug. 11, 2006 Research Highlight Biology Chemistry
How a fat cleaving enzyme works
Scientists solve the structure of an important fat cleaving enzyme and show how its ability to bind metal ions allows it to function
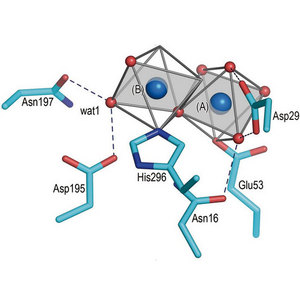 Figure 1: Bc-SMase in complex with a metal ion
Figure 1: Bc-SMase in complex with a metal ion
The fat molecule sphingomyelin is abundant in cell membranes throughout the body. Its breakdown by enzymes called sphingomyelinases is significant because one of its byproducts, ceramide, is an important signalling molecule in a variety of different cellular processes, as well as in many diseases including cancer and diabetes1.
In a report by scientists from the RIKEN SPring-8 Center in Harima and two other Japanese research institutes, the detailed atomic structure of one of these sphingomyelinases (Bc-SMase) has been determined—in this case from the bacterium Bacillus cereus. Knowledge gained from this structure should have direct application to humans because the enzyme in this bacterium has strong functional similarity to mammalian versions.
Bc-SMase can only work if it binds divalent ions such as Co2+ or Mg2+, and to a much lesser extent Ca2+. So to gain insight into the exact mechanism by which this enzyme binds these different ions and how they allow it to cleave sphingomyelin, Masashi Miyano and colleagues investigated at the atomic level the 3-dimensional structure of Bc-SMase bound to one of these three types of ions. Their findings are reported in The Journal of Biological Chemistry2.
The team identified the three different structures by separately crystallizing Bc-SMase in a complex with each type of ion, and then they shot a beam of X-rays at the isolated crystals using the SPring-8 synchrotron facility. From the diffraction pattern of the X-rays that scattered after hitting the crystals, it was possible to determine the different structures of the enzyme as it bound to each different ion (Fig. 1).
The structure revealed to the scientists exactly how ion binding allowed the enzyme to interact catalytically with the sphingomyelin. It also showed that it bound Ca2+ slightly differently from Co2+ and Mg2+, such that it was not able to as efficiently interact with the sphingomyelin, explaining why Ca2+ was a poorer co-factor for the enzyme. The research team also investigated a structurally unrelated portion of the protein from the ion-binding pocket and showed that it sits on the surface of the protein and allows it to bind cell membranes, which would bring it into proximity to the sphingomyelin.
Given the emerging importance of this class of enzymes, and its metabolic byproducts, in disease this structural information should go a long way in helping future research groups identify small molecules that can therapeutically target this class of enzymes.
References
- 1. Sawai, H, Domae, N, & Okazaki, T. Current status and perspectives in ceramide-targeting molecular medicine. Current Pharmaceutical Design 11, 2479–87 (2005). doi: 10.2174/1381612054367463
- 2. Ago, H., Oda, M., Takahashi, M., Tsuge, H., Ochi, S., Katunuma, N., Miyano, M., & Sakurai, J. Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus. The Journal of Biological Chemistry Epub, 4 April 2006 (PMID: 16595670). doi: 10.1074/jbc.M601089200
