Apr. 4, 2008 Research Highlight Biology
Controlling the message
New research reveals how cells prevent sensitive protein-encoding messages from falling into the wrong hands
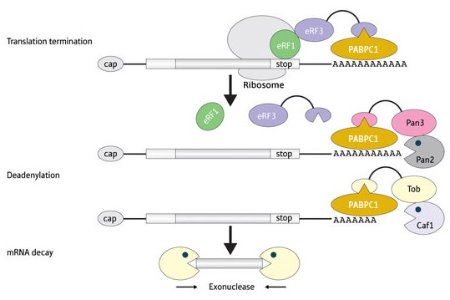 Figure 1: A schematic of the transition from mRNA translation to degradation.
Figure 1: A schematic of the transition from mRNA translation to degradation.
Like a spy receiving a coded secret message, cells typically destroy mRNA transcripts once they have been successfully translated into protein. As with the spy, this is for the cell’s own protection; the longer a transcript survives, the less control there is over the amount of protein generated from it.
This process requires precise coordination, and Yuji Funakoshi, a researcher in Masafumi Tsujimoto’s group at the RIKEN Discovery Research Institute in Wako, explains that “mRNA decay is intimately linked to and regulated by translation.” Until recently, however, the details of that linkage were unclear.
A key step in mRNA decay involves the removal of a long stretch of adenine nucleotides at the end of the transcript—a stabilizing structure known as the poly(A) tail—in a process called deadenylation. Previous work by Shin-ichi Hoshino’s group at Nagoya-city University has demonstrated that eRF3, a factor involved in translational termination, appears to mediate deadenylation via interaction with PABPC1, a protein that binds the poly(A) tail1. Now Funakoshi, Tsujimoto, and Hoshino have followed up on this work to examine how this interaction facilitates post-translational mRNA decay2.
In eukaryotic cells, mRNA degradation is managed by two specialized multi-protein complexes, Pan2–Pan3 and Caf1–Ccr4. By selectively disrupting the activity of these complexes in yeast and mammalian cells, Hoshino’s group demonstrated that eRF3 relies on both of them to trigger mRNA decay. This appears to be a two-stage process—Pan2–Pan3 causes slow but steady shortening of the tail, after which Caf1–Ccr4 induces rapid removal of the rest of the tail and subsequent transcript degradation.
Both complexes bind to PABPC1, which activates their enzymatic activity. Importantly, Pan2–Pan3 and Caf1–Ccr4 both interact with the same subdomain of PABPC1 as eRF3. This suggests a model where transcript decay is initiated when eRF3 recognizes that translation is complete and dissociates itself from PABPC1, freeing up this site for sequential binding by the two deadenylation complexes (Fig. 1).
“The most important finding in this research is that we have clarified the molecular mechanism that triggers mRNA decay,” says Funakoshi. “This mechanism plays a fundamental role in gene expression.” These findings also suggest other roles for eRF3, and the Hoshino group is now examining eRF3’s involvement with nonsense-mediated decay (NMD), a process that eliminates faulty mRNAs in which random mutations have introduced inappropriate ‘stop translation’ signals into a transcript. “eRF3 could play a pivotal role in NMD,” concludes Funakoshi.
References
- 1. Hoshino, S., Imai, M., Kobayashi, T., Uchida, N. & Katada, T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3’-Poly(A) tail of mRNA. Journal of Biological Chemistry 274, 16677–16680 (1999). doi: 10.1074/jbc.274.24.16677
- 2. Funakoshi, Y., Doi, Y., Hosoda, N., Uchida, N., Osawa, M., Shimada, I., Tsujimoto, M., Suzuki, T., Katada, T. & Hoshino, S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes & Development 21, 3135–3148 (2007). doi: 10.1101/gad.1597707
