Jul. 18, 2008 Research Highlight Biology
The giddy round of molecular timing
RIKEN synchrotron radiation sheds light on a dynamic reaction cycle
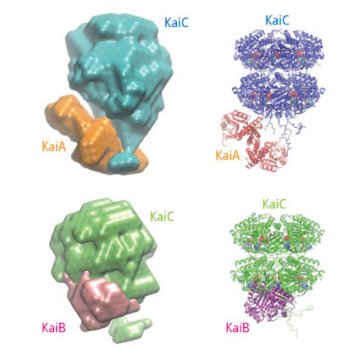 Figure 1: Low-resolution envelope models (left) and superimposed crystal structures (right) of the KaiA:KaiC complex (top) and the KaiB:KaiC complex (bottom). Reproduced with permission from Ref. 2 © 2008 Japan Synchrotron Radiation Research Institute (JASRI)
Figure 1: Low-resolution envelope models (left) and superimposed crystal structures (right) of the KaiA:KaiC complex (top) and the KaiB:KaiC complex (bottom). Reproduced with permission from Ref. 2 © 2008 Japan Synchrotron Radiation Research Institute (JASRI)
Japanese molecular biologists have determined the structures of a chronological sequence of complexes formed in an oscillating interaction among three proteins that acts as the timing mechanism for the circadian clock of blue-green algae, now known as cyanobacteria. This is the first time researchers have been able to observe such a biochemical system in motion.
Cyanobacteria are the simplest organisms known to have a biological clock. Earlier studies have shown the timing system relies on three clock proteins KaiA, KaiB and KaiC. When these three proteins are incubated with the phosphate-donor adenosine triphosphate, a stable cycle is established whereby KaiC gains and releases phosphate in reactions involving the other two Kai proteins.
Now, in a paper in Molecular Cell 1, researchers from the Japan Science and Technology Agency (JST), Nagoya University, and the RIKEN SPring-8 Center report they were able to use the powerful small-angle x-ray scattering (SAXS) beamline BL45XU at SPring-8 in Harima to follow the dynamic oscillatory reaction in real time. The intensity of forward scattering provides a sensitive measure of the average molecular weight of the compounds in solution and can be used to track the reaction cycle. The SAXS data could also be solved to provide low-resolution models of the complexes formed by the interactions of the proteins (Fig. 1).
Previous x-ray crystallographic studies determined that each of the proteins occurs as a multi-subunit structure, consisting of two subunits in the case of KaiA, four subunits in KaiB and a six-subunit barrel-shaped structure in KaiC. By determining the structures of the complexes formed when either KaiA or KaiB interact with KaiC, the researchers were able to show that the KaiC binding sites for the other two proteins were close enough for KaiA and KaiB to interfere with each other. KaiA binds to KaiC more quickly than KaiB, but the KaiB:KaiC interaction is thermodynamically more stable. During its interaction with KaiA phosphate is incorporated into KaiC, making the complex more attractive to KaiB which strips the phosphate away, making it more attractive to KaiA.
The oscillation cycle is driven initially by the assembly and disassembly of the KaiA:KaiC and KaiB:KaiC complexes, the researchers say, but subsequently is driven by the addition and removal of phosphate from KaiC. The researchers suggest two possible reaction cycles which fit their data. “The next target of our work is to determine which of our two possible schemes is more realistic,” says project-leader Shuji Akiyama of JST and RIKEN.
References
- 1. Akiyama, S., Nohara, A., Ito, K. & Maéda, Y. Assembly and disassembly dynamics of the cyanobacterial periodosome. Molecular Cell 29, 703–716 (2008). doi: 10.1016/j.molcel.2008.01.015
- 2. Akiyama, S., Nohara, A., Ito, K. & Maéda, Y. Real-time small-angle x-ray scattering observation of assembly and disassembly dynamics of cyanobacterial periodosome. SPring-8 Research Frontiers 2007 24–25 (2007). PDF version of the paper
