Sep. 4, 2009 Research Highlight Biology
Joining the dots on stem cell signaling
Hierarchical networks of transcription factors maintain self-renewal of mouse embryonic stem cells
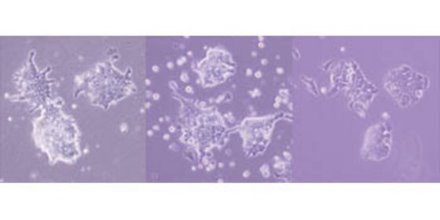 Figure 1: Examples of mouse embryonic stem cells grown without LIF, but with the transcription factors Klf4 (left), Nanog (middle) and TBx3 (right). Reproduced from Ref. 1 © 2009 Macmillan Publishers Ltd.
Figure 1: Examples of mouse embryonic stem cells grown without LIF, but with the transcription factors Klf4 (left), Nanog (middle) and TBx3 (right). Reproduced from Ref. 1 © 2009 Macmillan Publishers Ltd.
Transcription factors, the proteins that control the activity of genes, can be part of a hierarchy of signaling compounds, RIKEN molecular biologists have shown. They have also demonstrated such a hierarchy among the transcription factors and that they keep mouse embryonic stem cells from specializing or differentiating.
The study is important because the role of transcription factors in switching genes on and off is now recognized as a significant part of genetic function. For instance, researchers are now able to turn specialized cells back into a stem cell-like form—induced pluripotent stem cells—through applying transcription factors. Better understanding of how these factors themselves are activated should further this work.
Mouse embryonic stem cells in culture remain in an undifferentiated or pluripotent state if treated with the cytokine or extracellular hormone known as leukemia inhibitory factor (LIF). Inside the cell, such pluripotency is known to be directly associated with three transcription factors, Oct3/4, Sox2 and Nanog. In the past, other researchers have determined the involvement of the intermediate signaling compounds Jak and Stat3, and shown that pluripotency could be maintained without LIF by activating Nanog or Stat3 alone. How all these pieces fit together was unknown.
Hitoshi Niwa and colleagues from the RIKEN Center for Developmental Biology in Kobe set about tracing the signaling pathways, and detailed the results of their work in the journal Nature 1.
By analyzing data on compounds associated with the key transcription factor Oct3/4, they tracked down two other transcription factors, Klf4 and Tbx3. Either of these genes when artificially stimulated is capable, like Nanog, of maintaining pluripotency without LIF (Fig. 1). The researchers then created transgenic cells in which each of Klf4, Tbx3 and Nanog was activated, so they could study the impact of these transcription factors on levels of other key compounds.
Their work revealed parallel signaling pathways stimulated by LIF of a hierarchical nature. The pathway involving Jak and Stat3 turns out to activateKlf4 and through it Sox2 and Oct3/4. Tbx3 is part of another pathway which stimulates Nanog and Oct3/4. Other signaling compounds are known to connect into this latter pathway. They also found that transcription of all these factors is regulated by the core of Oct3/4, Sox2 and Nanog. The complexity of the network confers stability, the researchers say.
“Based on this picture, we will try to establish a precise quantitative model of the transcription factor network that will be applicable for computational simulation,” Niwa says.
References
- 1. Niwa, H., Ogawa, K., Shimosato, D. & Adachi, K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460, 118–122 (2009). doi: 10.1038/nature08113
