Nov. 4, 2011 Research Highlight Biology
Shedding light on a photosensitive protein
Special techniques for handling membrane proteins provide insight into the structure and function of an algal light-sensing molecule
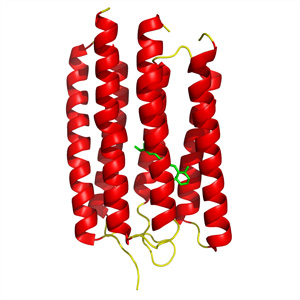 Figure 1: Crystal structure of ARII, a light-activated proton pump from the algaeAcetabularia acetabulum. © 2011 Shigeyuki Yokoyama
Figure 1: Crystal structure of ARII, a light-activated proton pump from the algaeAcetabularia acetabulum. © 2011 Shigeyuki Yokoyama
Even without eyes, many single-celled organisms can perceive and react to light. This is achieved via rhodopsins, proteins at the cell surface that trigger responses to specific wavelengths of light by directing the flow of ions into or out of the cell.
Naoki Kamo’s group at Matsuyama University in Ehime recently began working with ARII, a gene encoding a rhodopsin from the algae Acetabularia acetabulum. The encoded ARII protein proved extremely difficult to characterize and its function was initially ambiguous. However, Kamo’s team found success by joining forces with Shigeyuki Yokoyama’s group at the RIKEN Systems and Structural Biology Center in Yokohama1.
To reveal a protein’s structure and function, scientists typically generate highly ordered crystals of that protein and then analyze the diffraction pattern that results when the crystals are bombarded with x-rays. Membrane proteins will fold only under very specific conditions, but Yokoyama’s team devised a ‘cell-free’ system that provides tight control over protein manufacture2. By mixing the cellular protein synthesis machinery with lipids and detergents, they were able to achieve an environment highly hospitable to ARII production.
“This tough target could be expressed very efficiently using our cell-free protein synthesis system, even to the same degree as easy, soluble proteins,” says Yokoyama. He was subsequently able to rapidly purify the resulting protein and obtained a high-resolution structure for ARII by crystallizing it in the presence of lipid molecules (Fig. 1).
ARII proved to be relatively similar to bacteriorhodopsin (BR), a proton pump from the archaeal species Halobacterium salinarum. Preliminary analysis of ARII suggested that this protein likewise acts to transport protons from the cytoplasm to the exterior of the cell in response to illumination.
By analyzing the ARII structure, the researchers were able to identify a network of amino acids that directly participate in the uptake and release of individual protons. There are some notable differences in the kinetics of proton transport between BR and ARII. Kamo and Yokoyama also noted subtle structural disparities that might explain why ARII releases its protons ‘late’ relative to the rapid release observed with BR.
Having demonstrated the effectiveness of this membrane protein synthesis approach, the researchers are now delving deeper into the structure and function of ARII and ARI, another rhodopsin expressed by A. acetabulum. “We will produce various mutants with this efficient cell-free system and use many biophysical methods to understand the detailed proton transport mechanism and physiological roles of ARI and ARII,” says Yokoyama.
References
- 1. Wada, T., Shimono, K., Kikukawa, T., Hato, M., Shinya, N., Kim, S.Y., Kimura-Someya, T., Shioruzu, M., Tamogami, J., Miyauchi, S. et al. Crystal structure of the eukaryotic light-driven proton-pumping rhodopsin, Acetabularia rhodopsin II, from marine alga. Journal of Molecular Biology 411, 986–998 (2011). doi: 10.1016/j.jmb.2011.06.028
- 2. Shimono, K., Goto, M., Kikukawa, T., Miyauchi, S., Shirouzu, M., Kamo, N. & Yokoyama, S. Production of functional bacteriorhodopsin by an Eschericihia coli cell-free protein synthesis system supplemented with steroid detergent and lipid. Protein Science 18, 2160–2171 (2009). doi: 10.1002/pro.230
