Dec. 21, 2007 Research Highlight Biology Chemistry
Tackling cancer by fishing for trouble
Detecting sequence-specific problems in DNA may help scientists understand genetic disorders that lead to cancer
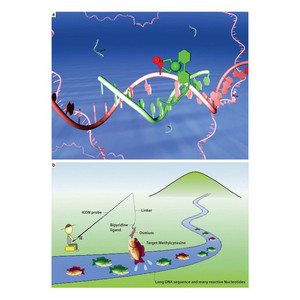 Figure 1: An illustration of how the complex between the osmium-containing DNA strand probe (green strand: fishing rod, hook and bait) would bind the target methylcytosine (red tag) in a biological sample of DNA (pink strand). (a) A biological model and (b) a cartoon of the technique.
Figure 1: An illustration of how the complex between the osmium-containing DNA strand probe (green strand: fishing rod, hook and bait) would bind the target methylcytosine (red tag) in a biological sample of DNA (pink strand). (a) A biological model and (b) a cartoon of the technique.
A team of Japanese scientists has developed a method to ‘fish’ for specific DNA-base problems that are responsible for causing cancer. The method can also be used to identify the exact site and measure the extent of the disorder.
Akimitsu Okamoto from the RIKEN Frontier Research System, Wako, has been leading this successful team for two years in developing this research aimed at detecting specific changes in DNA that cause ageing and may lead to the onset of cancer.
One of the bases in DNA is cytosine which, during normal biological processes, can react in our bodies by adding a methyl group and result in a compound known as methylcytosine. Whilst this reaction is important for gene regulation, excessive reaction has been shown to be carcinogenic; therefore detecting methylcytosine is of medical significance. Conventional methods exist to detect this target compound; however they all involve long reaction times and large samples—which are often destroyed—with little or no selectivity.
The latest technique by Okamoto and co-workers builds on their previous research to selectively detect methylcytosine, and now facilitates sequence-specific detection within DNA. The technique has reaction times of under an hour1. The advantage of a sequence-specific approach is that the exact origins of excessive methylation, and consequently carcinogenesis, can be investigated.
Using this conceptually new approach that employs techniques from interdisciplinary areas of organometallic chemistry and DNA physics, the researchers essentially fish for methylcytosine within a sample (Fig. 1). The detection probe, named ICON, is a short sequence of DNA that acts as the fishing rod. A ligand, made from bipyridine, acts as the fish-hook. The target methylated cytosine is then caught using a transition metal, osmium, as bait. In the sample, a complex is formed between the osmium, bipyridine and the methylcytosine—this is the fish on the hook. Unmethylated cytosine cannot form a complex with osmium, so the fishing bait is ignored.
The complex that is formed, a cross-linked structure between the two parts, is easily detected. This new technique clearly distinguishes not only between methylated and non-methylated cytosines, but also quantifies the degree and site of methylation.
Okamoto is confident this research will lead to further improvements and the development of a reliable method for methylcytosine detection. “The reaction conditions need to be optimized to establish the best analyzing system,” he explains. “The concept of crosslink formation would be applicable to DNA chips allowing analysis using familiar systems such as fluorescence detectors.”
References
- 1. Tanaka, K., Tainaka, K., Umemoto, T., Nomura, A. & Okamoto, A. An osmium–DNA interstrand complex: Application to facile DNA methylation analysis. Journal of the American Chemical Society 129, 14511–14517 (2007). doi: 10.1021/ja076140r
