May 2, 2014 Perspectives Biology
Seeing the big picture in cells
By identifying the common characteristics of molecular and cellular dynamics, researchers in quantitative biology are working toward models of whole cells and entire tissues.
Biology requires both bottom-up studies of molecular interactions and top-down approaches that combine these findings into larger networks. By identifying the common characteristics of molecular and cellular dynamics, researchers in quantitative biology are working toward models of whole cells and even entire tissues. These models—informed by data from advanced imaging techniques—act as virtual laboratories for exploring how biological systems respond to environmental changes and disease.
 © 2014 RIKEN
© 2014 RIKEN
The future of medicine, agriculture and biotechnology relies on scientists continually learning more about molecules and cells and their interactions. By necessity, many biological studies focus on individual components of these systems, such as pinpointing the roles of specific genes in the progression of diseases. However, the biological systems that determine life comprise many components that operate in complex and dynamic networks and are influenced by external environmental factors.
The challenge remains to develop new measuring, imaging and analytical techniques that reveal these networks and their workings. Incorporating this information into models that describe cells and tissues in their entirety will give researchers the capability to design artificial systems that predict the body’s response to drugs or show how to make cells behave in useful ways. These are the ambitious aims of scientists working in the expanding field of quantitative biology.
Although a seemingly overwhelming task, certain common themes have emerged, allowing generalized rules to be adopted within whole-system models. One dominant theory at the RIKEN Quantitative Biology Center (QBiC) is that dynamic biological networks are scale-invariant, which means that such rules apply from the molecular, subnanometer level up to the whole-tissue level.
Helpful heterogeneity
Within any dynamic biological network, there will be cells or molecules that are otherwise identical but react to external stimuli in very different ways. A given population of cells will contain a subset of individuals that thrive in the current environment and another subset of individuals that do not. Should the environment change, then the new conditions might favor the latter subset. This population characteristic is known as heterogeneity.
One famous demonstration of heterogeneity is the peppered moth, which exists with both white and black body colorings. As a consequence, different individuals are camouflaged against different tree barks. In a normal environment with white trees, the white moth prospers, but in the presence of heavy industry, the tree bark becomes darkened by pollution and the black-bodied moth is favored.
Similarly, at the subcellular level, there is heterogeneity in the states that molecules can adopt, allowing them to serve different functions. For example, signaling proteins within a cell respond to external stimuli by binding to nearby so-called effector molecules. Advanced single-molecule microscopy and spectroscopy studies by QBiC and others have verified that signaling molecules exist in several different structural states, which bind to different effectors in order to play different regulatory roles1.
The random nature of nature
Typically, heterogeneity emerges spontaneously through random noise. Hence, any realistic models must include relationships based on random, or stochastic, principles.
Stochastic processes are particularly important in the nanoscopic world. At these scales, movement is largely governed by Brownian motion—the random movement of molecules caused by collisions with surrounding particles. The cargo transporter molecule myosin, for example, has a two-headed structure that moves in a hand-over-hand fashion to climb along cytoskeletal filaments of actin. While such a movement might appear deterministic, single-molecule imaging has shown that when one head of the myosin molecule is free, it moves back and forth along the actin filament under Brownian motion. The head waves around until it chances upon the optimum position further up the filament, where it makes a strong bond and thus pulls its cargo to the targeted location in the cell2. These findings by QBiC researchers could have unexpected spin-off applications, such as informing the development of nanosized motors for miniature devices.
Seeing how cells break out
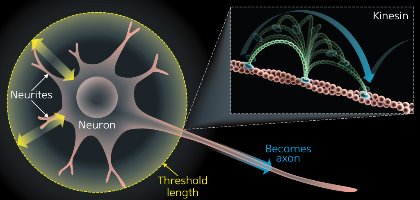 Figure 1: Observing Brownian motion with single-molecule imaging. Neurons start with multiple, identical neurites that are growing and shrinking (yellow arrows). Eventually, one neurite will pass the threshold length and not retract, continuing to grow into an axon (blue arrow). The rest become dendrites. Material needed to grow the axon is transported by kinesin, which, like other motor molecules including myosin, moves in a hand-over-hand fashion driven by Brownian motion (inset). © 2014 RIKEN
Figure 1: Observing Brownian motion with single-molecule imaging. Neurons start with multiple, identical neurites that are growing and shrinking (yellow arrows). Eventually, one neurite will pass the threshold length and not retract, continuing to grow into an axon (blue arrow). The rest become dendrites. Material needed to grow the axon is transported by kinesin, which, like other motor molecules including myosin, moves in a hand-over-hand fashion driven by Brownian motion (inset). © 2014 RIKEN
Heterogeneity and stochastic processes can also help explain how primitive cells change shape to become more specialized. All cells are symmetrical to begin with before they receive signals that initiate reorganization of their structure. This is well illustrated by neurons in the brain. Mature neurons have many protrusions, comprising one long axon and multiple shorter dendrites. Initially, these structures are identical, all undergoing a series of dynamic extensions and retractions. Eventually, one protrusion does not retract and continues to grow into an axon; all others become dendrites3 (Fig. 1). Single-molecule imaging studies have shown that the axon grows via stochastic processes through the accumulation of growth proteins.
This type of symmetry breaking can also be seen in whole-cell movement, or chemotaxis. This is the process by which a cell moves either toward or away from an area rich in chemicals, such as food or toxins. When signaling molecules within a cell pick up an external chemical signal, certain subcellular structures move from their original, random positions to assemble at a specific location, causing the cell to move. Research at QBiC has shown that cells can achieve the high sensitivity required for chemotaxis because their internal molecular components are in a constant state of flux4.
Bringing the whole cell together
By taking an integrated approach to analyzing the structural, spatial and temporal properties of cells, researchers at QBiC make up a vital part of the global effort to understand complex biological systems. Imaging techniques are vitally important for this work: through innovations in microscopy, molecular probes and other measurement systems, researchers hope to obtain simultaneous nanometer and millisecond resolutions for the three-dimensional imaging of molecules inside living cells.
Computational biologists have the daunting task of incorporating this information into models. Modeling protein interactions requires a timescale of milliseconds with a time resolution of femtoseconds (one quadrillionth of a second). For this reason, the future of whole-cell models will require researchers to exploit the full capabilities of supercomputers and develop algorithms based on the unifying principles described above.
Two computational tools lead this research at QBiC: MDGRAPE-4, a supercomputer designed to perform molecular dynamics simulations; and E-Cell, a software platform for modeling complex, heterogeneous and dynamic systems inside the cell. MDGRAPE-4 simulates the behavior of atoms and predicts how this will determine molecular behavior whereas E-Cell predicts how molecules operate at larger scales. Such comprehensive descriptions of activity at different scales will provide not only a better understanding of heterogeneous populations but also the potential to control them.
This control comes in the form of the third component of research at QBiC: synthetic design. RIKEN scientists are using data from imaging and computational work at QBiC to synthesize proteins without the need for a cell and to create lab-on-a-chip devices that can efficiently sort cells. These technologies are helping other researchers, for example, in creating strains of laboratory mice with specific traits that are useful for studying the effects of diseases and developing drugs.
At its core, QBiC is creating synthetic systems that act like biological ones. This work is possible thanks to a symbiotic relationship among researchers, from experimentalists to computational theorists to synthetic biologists. By continuing to uncover the operating principles that determine a cell’s state, researchers at QBiC are innovating technologies with the aim of controlling cell behavior.
References
- 1. Nygaard, R., Zou, Y., Dror, R. O., Mildorf, T. J., Arlow, D. H., Manglik, A., Pan, A. C., Liu, C. W., Fung, J. J., Bokoch, M. P. et al. The dynamic process of β2-adrenergic receptor activation. Cell 152, 532–542 (2013). doi: 10.1016/j.cell.2013.01.008
- 2. Iwaki, M., Iwane, A. H., Shimokawa, T., Cooke, R. & Yanagida, T. Brownian search-and-catch mechanism for myosin-VI steps. Nature Chemical Biology 5, 403–405 (2009). doi: 10.1038/nchembio.171
- 3. Fivaz, M., Bandara, S., Inoue, T. & Meyer, T. Robust neuronal symmetry breaking by Ras-triggered local positive feedback. Current Biology 18, 44–50 (2008). doi: 10.1016/j.cub.2007.11.051
- 4. Matsuoka, S., Shibata, T. & Ueda, M. Asymmetric PTEN distribution regulated by spatial heterogeneity in membrane-binding state transitions. PLoS Computational. Biology 9, e1002862 (2013). doi: 10.1371/journal.pcbi.1002862
About the Researcher
Toshio Yanagida

Recently honored as a Person of Cultural Merit by the Japanese government, Toshio Yanagida is a leading scientist in the area of the dynamics of biological systems. His research focuses on the development of new technologies, such as single-molecule imaging, for the study of biological molecular motors.
