Dec. 1, 2017 Research Highlight Biology
Molecular structure of elongation complex essential for DNA transcription determined
The newly revealed structure of a key complex involved in copying DNA will lead to a better understanding of the process
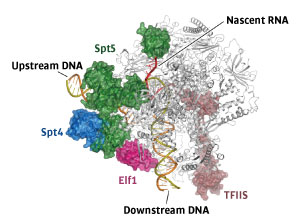 Figure 1: The elongation complex of polymerase II establishes well-defined paths for DNA and RNA. © 2017 From Ref 1. Reprinted with permission from AAAS.
Figure 1: The elongation complex of polymerase II establishes well-defined paths for DNA and RNA. © 2017 From Ref 1. Reprinted with permission from AAAS.
By using x-ray crystallography and cyro-electron microscopy data, an all-RIKEN team has elucidated the structure of a complex that plays a critical role in the copying of DNA into RNA1. This structural information provides vital clues as to how the complex functions.
Before genes encoded in DNA can be expressed, the relevant section of DNA has to be transcribed into RNA. It is simple to transcribe DNA in a test tube—all you need are DNA, RNA polymerase and nucleotides—but the situation is more complex in a cell’s nucleus. “Many components, including transcription factors, associate with RNA polymerase II and form huge transcription complexes, many of which have not been structurally explored yet,” notes Shun-ichi Sekine from the RIKEN Center for Life Science Technologies.
His team is working on determining the structure of these large transcriptional complexes. In the current study, they focused on the elongation complex that forms in the early steps of transcription. It both ensures that transcription proceeds and regulates other essential processes linked to transcription, such as DNA repair. Sekine believes it is crucial to gain a full understanding of the complex’s mechanism in order to ascertain how this regulation can fail in diseases such as cancer.
 Shun-ichi Sekine of the RIKEN Center for Life Science Technologies and his team have determined the precise molecular structure of the elongation complex essential for DNA transcription. © 2017 RIKEN
Shun-ichi Sekine of the RIKEN Center for Life Science Technologies and his team have determined the precise molecular structure of the elongation complex essential for DNA transcription. © 2017 RIKEN
His team focused on three proteins known to be essential for the elongation complex: the multidomain protein Spt5, its binding partner Spt4 and the elongation factor Elf1. They determined the domains that are important for the interaction between Spt5 and the RNA polymerase. The researchers then solved the crystal structure of one Spt5 domain, called KOW5, complexed with the polymerase and Elf1. They also obtained a model of the entire complex by merging this crystal structure with cryo-electron microscopy data of a complex that included Spt4, Spt5 and a transcription factor.
These results provide information on how the complex operates. “We found that the elongation factors Spt4/5 and Elf1 bind to functionally important parts of polymerase II,” says Sekine, adding that they act like subunits of the polymerase.
Sekine likens the polymerase’s structure to a needle, where DNA is a thread that can pass through the needle’s eye. Elf1 forms an entry tunnel that helps thread the DNA into the polymerase and keep it in place. Spt4 and 5 make up an exit tunnel that separates DNA from RNA and ensures efficient exit of DNA and nascent RNA from the complex (Fig. 1). This aspect is critical for avoiding DNA–RNA hybridization, which can prematurely terminate transcription.
“Our structure provides a firm foundation for further structural, molecular and cellular biology studies and research on disease mechanisms,” says Sekine.
Related contents
- How DNA traffic jams cause cell differences
- Insights into the geometry of genetic coding
- Translating DNA into protein
References
- 1. Ehara, H., Yokoyama, T., Shigematsu, H., Yokoyama, S., Shirouzu, M. & Sekine, S. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science 357, 921–924 (2017). doi: 10.1126/science.aan8552
