Jul. 26, 2019 Research Highlight Biology
A new model for exploring cell polarity
A method for inducing polarization in cells will allow scientists to explore the real-time processes that occur during cell polarization
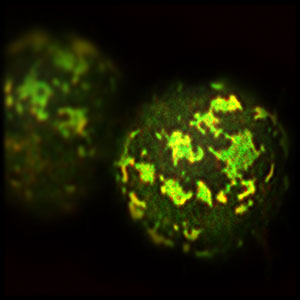 Figure 1: A super-resolution micrograph of artificially polarized Schneider cells, or S2 cells. © 2019 RIKEN Center for Biosystems Dynamics Research
Figure 1: A super-resolution micrograph of artificially polarized Schneider cells, or S2 cells. © 2019 RIKEN Center for Biosystems Dynamics Research
RIKEN researchers have found an intrinsic way to induce cell polarity—the asymmetry observed in the shape, structure, or organization of cells—in fruit fly cells1. This method provides a useful means for studying the dynamics of cell polarization on a micrometer scale.
Animals consist of many different types of cells, all of which originate from a single cell—the fertilized egg. To generate this huge diversity in cells, the egg and its descendants must divide unevenly to produce new cells with different fates. Thus, asymmetric cell division, which generates two different daughter cells, is a critical step in cell development. Cell polarity plays a crucial role in asymmetric division.
Much work has been done on discovering the factors that drive cell polarity. In particular, the Par family of proteins has been found to play a crucial role in establishing polarity.
“As our understanding of cell polarity advanced, we noticed that the Par complex operates in a context-specific way,” says Fumio Matsuzaki of the RIKEN Center for Biosystems Dynamics Research. “This has made it difficult to study the general features of the cell polarization process.”
To gain a better picture of polarization dynamics, the team focused on generating Par-dependent polarity using non-polar cells.
The idea behind this approach had been simmering in Matsuzaki’s mind for more than a decade, but work did not begin in earnest until graduate student Kalyn Kono took on this challenging project 4 years ago. By investigating changes in the expression level of each of the major components of the Par complex, the team hit on their method for inducing polarity through overexpressing Par3—one of the three core proteins of the Par family—in non-polar fruit fly cells called Schneider 2 cells, or S2 cells (Fig. 1).
Using their reconstruction system, the researchers observed clusters, or what they call Par-complex islands, that are composed of a meshwork containing unit-like segments, which dynamically associated and dissociated with each other. As these island structures resemble patch-like features known to exist in the neural stem cells of fruit flies, their method appears to closely model the autonomous process of polarization.
“Our success in polarizing S2 cells implies that, in principle, any cell can be polarized if a certain set of conditions is met,” comments Matsuzaki.
A remaining question is why the Par-complex islands do not appear to merge into one large island under the cell membrane, even when polarized. “We have started collaborating with physicists and biophysicists to tackle this interesting problem,” Matsuzaki adds.
The team intends to explore what determines whether the Par-complex islands cluster asymmetrically or evenly. Going beyond polarization, a future goal is to reconstruct asymmetric cell division.
References
- 1. Kono, K., Yoshiura, S., Fujita, I., Okada, Y., Shitamukai, A., Shibata, T. & Matsuzaki, F. Reconstruction of Par-dependent polarity in apolar cells reveals a dynamic process of cortical polarization. eLife 8, e45559 (2019). doi: 10.7554/eLife.45559
