Mar. 16, 2007 Research Highlight Biology
Proteins pumping protons
Researchers unlock the secret of an important enzyme
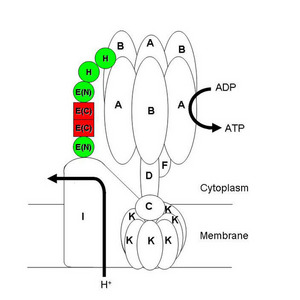 Figure 1: A model of the stator part of A-ATPase is shown with its dimeric core (red) and a modular architecture (red and green). J. Mol. Biol./ Elsevier/ 366/ 941 (2007).
Figure 1: A model of the stator part of A-ATPase is shown with its dimeric core (red) and a modular architecture (red and green). J. Mol. Biol./ Elsevier/ 366/ 941 (2007).
A RIKEN research team has uncovered significant details of the internal structure of a protein complex which actively transports protons across membranes in cells.
The complex on which the team worked is an H+-ATPase found in primitive, single-celled organisms known as archaea. But it is closely related to a similar enzyme of the internal cell membranes in higher animals, where it is responsible for maintaining the acidic environment required for many cellular processes, and plays an important role in the spread of cancer and in bone resorption, hence osteoporosis.
H+-ATPases use the energy released in breaking the chemical bond to the terminal phosphate in the universal energy molecule adenosine triphosphate (ATP) to power the transfer of a proton (H+) across a membrane. They are constructed out of many different protein-chain subunits—nine in the archaeal version (A-ATPase) studied. The proton transport part of the complex spans the membrane, and the ATP-ase is attached to it by means of a protein spike, known as a stator (Fig. 1). It is through this stator that the energy of ATP-breakdown is linked to proton transport. Until now, neither the structure, nor even the numbers of stators per A-ATPase complex were known.
From previous work, the heart of the stator was known to be a protein chain called subunit E. In a recent paper in the Journal of Molecular Biology1, researchers from the RIKEN SPring-8 Center in Harima reported the first crystal structure of subunit E. They also studied the relationship of subunit E with another stator protein chain, subunit H, using electrophoresis, mass spectrometry, amino acid sequencing and circular dichroism spectroscopy, which measures the coiling of proteins.
They found that the ends of two subunit E chains bind together to form a two-part or dimeric working core of the stator structure (Fig. 1). The two other ends of subunit E bond either with subunit H or directly to the proton transfer part of the complex. The group suggests that subunit E, subunit H, and at least two other subunits in other H+-ATPases fit together like modular building blocks, and can be used to make stators longer or shorter to match the other structures of the complex.
“We now want to clarify the situation with other H+-ATPases, as they are all related evolutionarily,” says research team leader, Naoki Kunishima.
References
- 1. Lokanath, N.K., Matsuura, Y., Kuroishi, C., Takahashi, N. & Kunishima, N. Dimeric core structure of modular stator subunit E of archaeal H+-ATPase. Journal of Molecular Biology 336, 933–944 (2007). 10.1016/j.jmb.2006.11.088
