May 2, 2007 Research Highlight Biology
Insights into regulation of tumor suppression
Recent understanding of a protein structure could lead to new cancer drugs
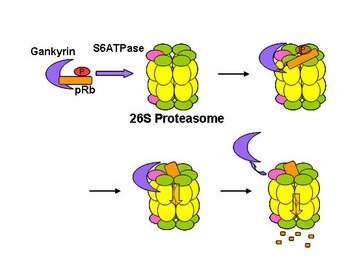 Figure 1: The proposed model of gankyrin’s role in the degradation of the tumor suppressing protein pRb.
Figure 1: The proposed model of gankyrin’s role in the degradation of the tumor suppressing protein pRb.
Researchers from RIKEN have unraveled details of the interaction between an oncoprotein, involved in regulating cancer, and the 26S proteasome, which breaks down proteins. The complex they form is part of the degradation pathway of at least two important tumor suppressing proteins, p53 and retinoblastoma protein (pRb). The work has suggested a mechanism for the breakdown of the suppressors, and could lead to the design and development of new drugs to treat cancer.
The oncoprotein, gankyrin, is commonly associated with liver cancer. Earlier research has shown that it interacts directly with pRb and with components responsible for pRb’s activation and degradation. Gankyrin is also involved in tagging the ubiquitous tumor suppressor p53 for degradation. It was first identified, however, as binding with a regulatory particle which unfolds and feeds specially tagged proteins into the barrel-shaped 26S proteasome where they are chopped up and destroyed.
Although gankyrin is known to associate with S6 ATPase, one of six different ATPase enzymes that are part of the regulatory particle, the details of this interaction have never been clear. In a recent paper in Structure1, researchers from the RIKEN Genomic Sciences Center in Yokohama provide the first details of the gankyrin—S6 ATPase complex. They were obtained by means of x-ray crystallography using synchrotron radiation.
The structure shows how a concave region of gankyrin binds strongly to complementary electrostatic charges in a region close to one end of S6 ATPase. Mutations which change the charge distribution in this region of S6 ATPase result in a dramatic loss of binding affinity for gankyrin. And gankyrin does not bind to any of the other ATPases in the regulatory particle, presumably because they do not have the correct charge distribution.
The researchers also found that gankyrin could interact with S6 ATPase whether it was attached to the proteasome or not. Gankyrin also interacts with pRb, whether associated with S6 ATPase or not. It does not bind as strongly to pRb, however, as to the ATPase. The suggestion from all this evidence is that gankyrin can function to carry phosphorylated pRb to the 26S proteasome where it binds strongly to S6 ATPase. Subsequently, gankyrin releases pRb for destruction in the proteasome (Fig. 1).
“We want to understand how this protein regulates cancer through interaction with other proteins,” say research team members, Balasundaram Padmanabhan and Shigeyuki Yokoyama. “This is an expanding project. So far, only this one complex structure is known.”
References
- 1. Nakamura, Y., Nakano, K., Umehara, T., Kimura, M., Hayashizaki, Y., Tanaka, A., Horikoshi, M., Padmanabhan, B. & Yokoyama, S. Structure of the oncoprotein gankyrin in complex with S6 ATPase of the 26S proteasome. Structure 15, 179–189 (2007). doi: 10.1016/j.str.2006.11.015
