Jan. 4, 2008 Research Highlight Biology
Clockwork orange beats time for the body
Researchers find new master gene setting circadian rhythms

A RIKEN-led team of researchers from Japan and the US has used an innovative combination of genome survey techniques in live Drosophila fruit flies to reveal a previously unknown master gene involved in setting circadian rhythms. It is the tenth of a series of genes which generate proteins that interact in complex interlocking feedback loops to measure the 24-hour day.
This network of genes ensures that the rhythms of vertebrate animals—sleep and wakefulness, changes in body temperature and blood pressure, the secretion of hormones and regulation of fertility—are attuned to daily and seasonal cycles. The importance of the network’s role is demonstrated not only by the fact that mutations in clock genes have been linked to cancer and obesity, but also that they have been highly conserved during evolution and are similar in organisms from fruit flies to mammals. The regulation of the network is intricate and complex because, in addition to the simple measurement of time, it has to respond to changing daily patterns of light and dark and seasonal patterns of temperature and the environment.
In humans, such common problems as jet lag and lack of alertness of shiftworkers arise when the body’s circadian rhythms are not properly adjusted to the external environment. Permanent disruption of the body’s clock can lead to much more serious disorders, such as delayed sleep phase syndrome, and has also been implicated in the depressive mental illness known as seasonal affective disorder (SAD). Work on clock genes could well have relevance to treating these conditions.
Revealing the master gene
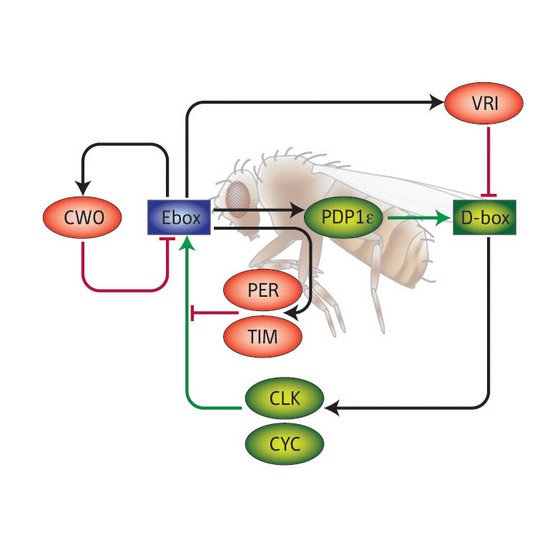 Figure 1: Schematic diagram of the relationships of the proteins now thought to be involved in the Drosophila circadian clock. Activators are in red, repressors in green.
Figure 1: Schematic diagram of the relationships of the proteins now thought to be involved in the Drosophila circadian clock. Activators are in red, repressors in green.
In a recent issue of the journal Genes & Development 1, the research group—from RIKEN’s Center for Developmental Biology in Kobe, Kyushu University, Japan’s National Institute of Genetics and two universities in Texas—detailed how it found the new master gene. The gene exhibits rhythmic daily activity and codes for a protein that represses other clock genes through the regulatory DNA sequence known as E-box. Because it carries a characteristic DNA domain known as ORANGE, the research team named the gene clockwork orange (cwo ) referring to the well-known Anthony Burgess novel and Stanley Kubrick film.
The researchers uncovered cwo using a novel partnering of micro-array technology that shows which genes are cyclically switched on, with RNA interference techniques, whereby individual genes can be switched off at will. In previous work, using micro-arrays they had followed the daily activity pattern of all genes in the Drosophila genome in cells of the head region of the fruit fly, where the circadian pacemaker is located. They found about 200 genes showed a rhythmic activity pattern over the daily cycle both under normal conditions of light and dark, and also in constant darkness.
In the recently reported study, using RNA interference techniques the researchers then switched off each of about 130 genes which showed such regular daily cycles of activity. They were looking for dramatic disturbances of overall rhythmic behavior which, they hypothesized, would only happen when they disrupted the core genes involved in setting the body’s clock.
In addition to genes already known to be an integral part of the circadian mechanism in Drosophila, the researchers found five other candidates for such clock genes. Of these, cwo was the gene with the most pronounced and stable impact.
Understanding the mechanism
Using a combination of micro-array technology and antibodies, the group then set out to discover the genes with which cwo interacts. They found that the genes to which the protein product CWO binds tend to contain the E-box DNA sequence. These included several genes known to play a key role in the network that regulates the body’s clock, as well as cwo itself. CWO represses the activity of all the genes to which it binds. Interestingly, the overall effect of lowering CWO activity is to dampen the amplitude of the circadian rhythm.
The core molecular feedback system which establishes the daily biological clock in Drosophila is thought to involve four proteins known as CLK (clock), CYC (cycle), PER (period) and TIM (timeless) and their linked genes (Fig 1). CLK and CYC bind together and stimulate the production of PER and TIM by forming a complex with E-box proteins. As PER and TIM begin to accumulate, they feed back to inactivate the CLK–CYC complex. Several other proteins interact with this basic feedback loop, regulating and fine-tuning the mechanism.
The picture which begins to emerge from the latest work is one where CWO regulates this basic mechanism by competing with the CLK–CYC complex to bind to the E-box proteins. CWO also regulates itself by binding to and repressing the transcriptional activity of the cwo gene.
“The work is still far from complete,” says Hiroki Ueda, the research team leader. “But I feel the discovery of cwo, which has a counterpart in the human genome, represents an important step in deciphering biological clocks. We next want to apply our techniques to the mouse, which is very near to humans compared with the fruit fly.”
References
- 1. Matsumoto, A., Ukai-Tadenuma, M., Yamada, R.G., Houl, J., Uno, K.D., Kasukawa, T., Dauwalder, B., Itoh, T.Q., Takahashi, K., Ueda, R., Hardin, P., Tanimura, T. & Ueda, H.R. A functional genomics strategy reveals clockwork orangeas a transcriptional regulator in the Drosophila circadian clock. Genes & Development 21, 1687–1700 (2007). doi: 10.1101/gad.1552207
About the Researcher
Hiroki R. Ueda

Hiroki R. Ueda was born in Fukuoka, Japan, in 1975. He graduated from the Faculty of Medicine, the University of Tokyo, in 2000, and obtained his PhD in 2004 from the same university. During his undergraduate studies he worked as a research assistant on a biological simulation system project at Sony Computer Science Laboratories. During his graduate studies from 2000, he went on to work as a researcher, and in 2002 became group leader at Yamanouchi Pharmaceutical, on a project studying biological-clock mechanisms in flies and mice. He was appointed laboratory head at the RIKEN Center for Developmental Biology (CDB) in April 2003 and manager of the Functional Genomics Subunit at the CDB from October 2004. He also became a visiting professor in Tohoku University between April 2005 and March 2006, and Tokushima University from April 2005, and has been an invited professor at Osaka University since April 2006. His research interests include system-level understanding of biological time, space and information, and systems-based medicine on human disease.
