May 16, 2008 Research Highlight Biology
No loose ends
Tying short RNA molecules into loops gives them a stability boost, which could lead to more effective therapeutic strategies for modulating gene expression
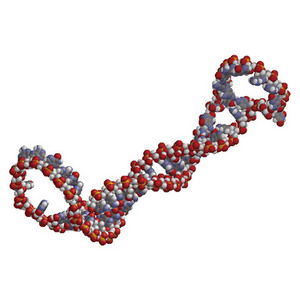 Figure 1: Three-dimensional structure of one of the Ito group’s ‘dumbbell siRNA’ constructs.
Figure 1: Three-dimensional structure of one of the Ito group’s ‘dumbbell siRNA’ constructs.
RNA interference, in which short interfering RNA (siRNA) molecules are used to target specific genes for downregulation in vivo, is among the most powerful molecular biology techniques to emerge in recent years. RNAi was developed as a research tool following the discovery of naturally occurring siRNAs, which are processed from larger RNA transcripts by the cellular enzyme Dicer to yield very small (typically 21-nucleotide) RNAs containing specific sequences complementary to their endogenous target genes.
Synthetic siRNAs can be expressed directly from transgenes as small, hairpin-forming RNAs, which are subsequently converted into linear molecules by the action of Dicer; alternatively, they can also be introduced directly into cells in a pre-linearized form. In the laboratory, siRNAs have delivered considerable precision and efficiency in modulating gene expression in cultured cells and model organisms. Many scientists also see considerable promise in clinical applications of this technology, although there are a number of serious technical roadblocks that remain to be overcome.
Delivering the message
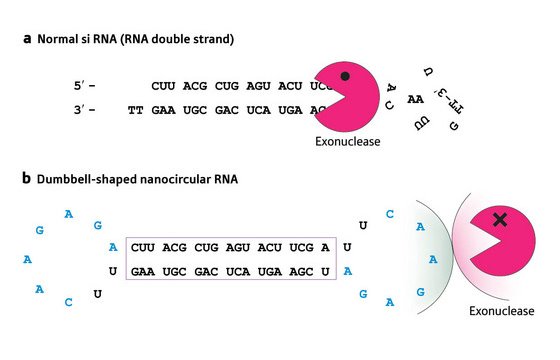 Figure 2: Linear siRNA molecules (a) are vulnerable to rapid degradation by exonuclease enzymes, which target the loose ends. Circularizing those ends (b) provides protection against exonuclease digestion, resulting in increased efficiency of inhibition of target genes.
Figure 2: Linear siRNA molecules (a) are vulnerable to rapid degradation by exonuclease enzymes, which target the loose ends. Circularizing those ends (b) provides protection against exonuclease digestion, resulting in increased efficiency of inhibition of target genes.
Chief among these impediments is finding a safe and effective means for siRNA delivery. Simple injection of siRNAs is not an option, as unprotected RNA molecules are rapidly degraded in the body, and so more complicated strategies are required—each with its own issues.
The introduction of fully functional siRNA genes into patients is also problematic, due largely to the same complications that plague gene therapy research. “Virus vectors that proliferate in vivo are potentially risky… vector systems can not be controlled in cells, and the dose of RNA is very important for clinical RNA interference,” explains Hiroshi Abe, a research scientist in Yoshihiro Ito’s laboratory at the RIKEN Advanced Science Institute in Wako. “If excess RNA is administered, cells will respond to these as foreign bodies, using the immune system.”
Another alternative involves the use of RNAs composed of unnatural nucleotides containing various chemical modifications. These synthetic RNAs are able to resist enzymatic degradation, and can therefore survive longer within the body, but Abe points out that this stability comes at the cost of reduced efficacy at gene silencing. “This remains a problem for unnatural molecules in RNAi technology,” he says.
Ito’s group has taken an innovative approach to deliver natural siRNAs effectively, based on the understanding that RNA-degrading enzymes typically start by chewing at loose RNA ends—they circularized their siRNAs, designing molecules that self-assemble into stable ‘dumbbell’ shapes1, with a base-paired central stem connecting two loops at either end (Fig. 1). By closing off the ends, the exonuclease should be left with no means of attack and the siRNA should acquire enhanced stability (Fig. 2).
Experimentally, these constructs behaved as expected, with the modified structure providing a considerable stability boost. Dumbbell and linear siRNA constructs were each incubated in the presence of exonucleases; after two hours, the percentage of intact linear siRNA was one-sixth that of the dumbbell construct. Similar results were obtained with RNAs incubated in human serum, which more closely mirrors the conditions likely to be experienced by siRNAs in clinical applications.
However, since naturally occurring siRNAs must also be processed by the enzyme Dicer before they can be effective, a key concern of Ito’s team was ensuring that their dumbbell constructs were not just stable, but also capable of being cleaved by Dicer and successfully inhibiting target genes.
The researchers treated a variety of linear and dumbbell constructs of different lengths with recombinant Dicer in order to examine processing efficiency, and found that the dumbbell constructs were generally capable of being processed, but typically at a far slower rate than the linear constructs.
This delayed processing did not impede their performance as gene inhibitors, however, and the dumbbell constructs generally surpassed their linear counterparts in triggering specific inhibition of target genes when injected into cultured human fibroblast cells. For some constructs, the improvement of suppression was as high as three-fold for the dumbbell siRNAs. The duration of effective gene suppression was also extended for the dumbbell constructs, perhaps as a result of their delayed processing by Dicer.
Building smarter dumbbells
Encouraged by these initial findings, Ito, Abe and colleagues have pursued strategies to further enhance the effectiveness of their constructs. Abe indicates that optimizing the dumbbell’s loop structure has become a major focus since the publication of this work, in order to develop optimized constructs that strike an effective balance between greater stabilization, efficient enzymatic processing, and potent inhibition of target genes.
“We have tried modification of the loop position and found that this offers improved stability,” says Abe. “We have also found that this loop is very important for controlling the editing position used by Dicer—in other words, we can decide the cleavage site of RNA by designing the loop position to produce appropriate siRNA species.”
With this progress in optimizing the biological activity of their dumbbell constructs, improving the means of delivery is now an important goal, and Abe adds that he and his colleagues are currently exploring the use of a variety of promising nanomaterials that could help deliver their dumbbell constructs into cells safely and efficiently.
References
- 1. Abe, N., Abe, H. & Ito, Y. Dumbbell-shaped nanocircular RNAs for RNA interference. Journal of the American Chemical Society 129, 15108–15109 (2007). doi: 10.1021/ja0754453
About the Researcher
Yoshihiro Ito

Yoshihiro Ito was born in Gifu, January 12, 1959. He received his Bachelor’s (1981) and Master’s (1983) degrees in polymer chemistry at Kyoto University and was awarded a doctorate in engineering from the same university in 1987. Since then he has held a number of posts at various institutions including Research Fellow of the Japan Society for the Promotion of Science (1987), assistant (1988) and associate (1998) professor at Kyoto University, research fellow at the University of California, Irvine (1992–1993), professor of the University of Tokushima (1999), and Project Leader at the Kanagawa Academy of Science and Technology (2002–2007). He has also been a visiting professor at Zhengzhou University, the Nagasaki Institute of Applied Sciences, Tokyo Metropolitan University and Tokyo Institute of Technology. Now, he is Chief Scientist and Director of the Nano Medical Engineering Laboratory at the RIKEN Institute of Physical and Chemical Research. His research focuses on biomaterial science, regenerative medical engineering, combinatorial bioengineering for the creation of functional polymers, and soft nanotechnology.
