Mar. 21, 2008 Research Highlight Biology
A cure for the cold
A recent structural biology study reveals how one protein helps keep bacteria running through a cold spell
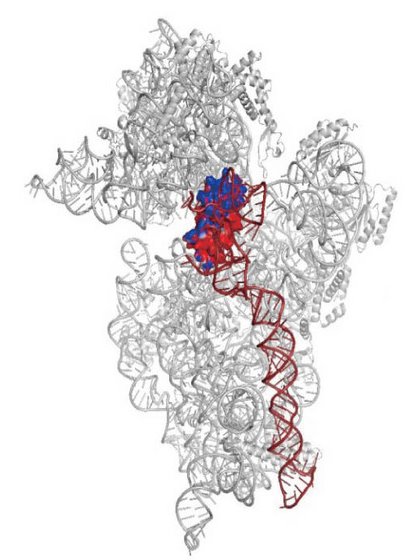 Figure 1: The structure of the RbfA-30S complex, revealing the interaction of the RbfA (red and blue) with helices (brown) on 30S involved in ribosomal assembly and RNA translation.
Figure 1: The structure of the RbfA-30S complex, revealing the interaction of the RbfA (red and blue) with helices (brown) on 30S involved in ribosomal assembly and RNA translation.
The translation of RNA into protein is managed by cellular machines known as ribosomes. A ribosome consists of two subunits, each composed of an RNA-based scaffolding with a number of specialized proteins attached.
The formation and assembly of ribosomes within a living cell seems to be a complicated process, which can be strongly affected by environmental conditions. For example, extreme cold has a very negative impact on bacterial protein production. “Cold shock results in an increase in the level of non-translating ribosomes, and produces a temporary cessation of bacterial growth,” explains Shigeyuki Yokoyama of the Genomic Sciences Center in Yokohama. “Growth is then restored through the action of a set of cold-shock response proteins.”
Previous studies have implicated the bacterial protein RbfA as an important component of this cold-shock response, and Yokoyama’s team recently joined up with German and American scientists to explore the RbfA-ribosome interaction in order to better understand the mechanism of this process1. RbfA binds to the smaller 30S subunit of the bacterial ribosome, and so the researchers began by acquiring detailed structural data for the subunit alone and bound to RbfA.
These data showed that RbfA binds in the immediate vicinity of helix h1, a specific domain of 30S that is known to fold improperly under cold-shock conditions. They also revealed a dramatic conformational change in 30S after RbfA binding, affecting a separate ribosomal domain directly involved in the RNA decoding process as well as assembly of the large and small ribosomal subunits (Fig. 1). Subsequent comparison of the RbfA binding site on the ribosome against the binding sites used by other proteins associated with 30S subunit maturation indicated that these various proteins interact with many of the same ribosomal domains, suggesting mechanistic overlap.
Collectively, these results indicate that the binding of RbfA to 30S serves a dual purpose: enabling proper formation of the ribosomal subunit, as well as ensuring that mRNA transcripts do not associate with premature, incompletely folded subunits. The findings from this work should be broadly applicable with regard to understanding ribosomal maturation, for although RbfA production is markedly increased at temperatures where ribosome formation becomes inefficient, the protein is generally present at low levels under other cellular conditions. “Our results not only provide insight into the role of RbfA during maturation of the 30S subunit,” says Yokoyama, “but they also suggest how RbfA confers a translational advantage to cells under conditions of cold shock.”
References
- 1. Datta, P.P., Wilson, D.N., Kawazoe, M., Swami, N.K., Kaminishi, T., Sharma, M.R., Booth, T.M., Takemoto, C., Fucini, P., Yokoyama, S. & Agrawal, R.K. Structural aspects of RbfA action during small ribosomal subunit assembly. Molecular Cell 28, 434–445 (2007). doi: 10.1016/j.molcel.2007.08.026
