Dec. 12, 2008 Research Highlight Biology
From genes to plant metabolism
An analytical survey maps out the genetic connections of the flavonols
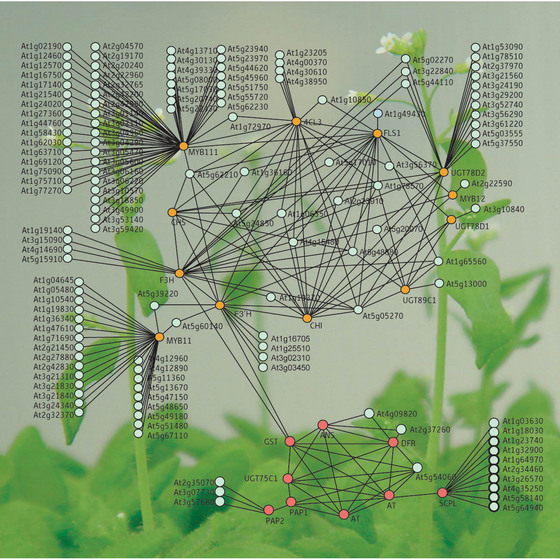 Figure 1: Relationships of the coexpression of genes involved in the flavonoid pathways in Arabidopsis. Orange circles represent known flavonoid-related genes, red circles represent the closely linked anthocyanin-related genes, and pale green circles represent the genes that may be involved in flavonoid metabolism.
Figure 1: Relationships of the coexpression of genes involved in the flavonoid pathways in Arabidopsis. Orange circles represent known flavonoid-related genes, red circles represent the closely linked anthocyanin-related genes, and pale green circles represent the genes that may be involved in flavonoid metabolism.
Plant molecular biologists from RIKEN’s Plant Science Center in Yokohama and Chiba University are using a combination of bioinformatics and biochemical analytical techniques to complete a map of all the reactions involving flavonols in the model plant Arabidopsis thaliana. Already the researchers have succeeded in identifying a new gene that encodes an important enzyme and in determining the physiological role played by another gene. The information gained in the study should be applicable to other plants.
Flavonols are health-promoting, dietary antioxidants. In plants, they are involved in defense responses, such as reactions to pathogens and UV radiation. The genes and biochemical pathways for flavonol synthesis have been well studied in several plants, but the details of subsequent chemical modification are less well known.
The research team is involved in a major project applying the latest techniques to finding the links between the genes of Arabidopsis, for which the entire genome has already been sequenced, and the compounds found in the plants themselves. The flavonol study is aimed at identifying these connections for a whole class of compounds, and results were published recently in The Plant Cell 1.
Initially, the researchers catalogued all the flavonol related compounds in Arabidopsis by comparing what was present in normal plants with what they found in a mutant form in which no flavonol was produced. They determined the compounds in the flowers, leaves, stems and roots of the plant by preparing extracts and putting them through liquid chromatography–mass spectroscopy (LC-MS) analysis. The study detected 30 flavonol-related products in Arabidopsis, some of which were intermediates for making others. Two other compounds have been reported in earlier studies.
Using plants with flavonol-related mutations, the LC-MS results, and data from previous studies, the researchers determined the structures and metabolic relationships of 15 newly identified and eight known flavonols. In a technique known as transcriptome coexpression analysis, they were then able to use software to identify the flavonol-related genes in the published genome, linking enzymes and regulatory factors with the products found in the plants themselves (Fig. 1).
On the basis of their results they undertook detailed analysis of two genes using genetically engineered mutants, uncovering one previously unknown gene and determining the metabolic role of another. “We now wish to complete our map of the flavonoids in Arabidopsis and then in other plant species, including the metabolic relationships,” says lead author, Keiko Yonekura-Sakakibara.
References
- 1. Yonekura-Sakakibara, K., Tohge, T., Matsuda, F., Nakabayashi, R., Takayama, H., Niida, R., Watanabe-Takahashi, A., Inoue, E. & Saito, K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene–metabolite correlations in Arabidopsis. The Plant Cell 20, 2160–2176 (2008). doi: 10.1105/tpc.108.058040
