Feb. 6, 2009 Research Highlight Biology
Neutralizing spurious associations
Statistical analysis helps the search for fundamental causes of disease
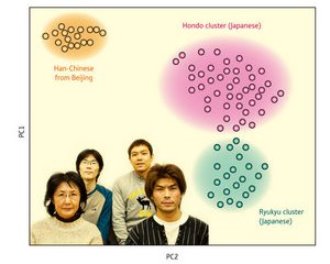 Figure 1: Using the genotype data for 140,387 SNPs in 7003 Japanese individuals, it was found that most of Japanese people fall into one of two distinct groups.
Figure 1: Using the genotype data for 140,387 SNPs in 7003 Japanese individuals, it was found that most of Japanese people fall into one of two distinct groups.
Most Japanese people fall into one of two distinct groups genetically, biostatisticians from RIKEN’s Center for Genomic Medicine in Yokohama have shown. The finding is significant because such population structure can bias genome-wide association studies (GWASs) aimed at determining genetic links to disease, leading to spurious associations. On the basis of their work the researchers suggest ways of avoiding or correcting this bias.
GWASs are increasingly employed to reveal relationships between single nucleotide polymorphisms (SNPs) and particular disease conditions. The technique seeks out statistical differences among sets of polymorphic genetic markers between sufferers of a disease and a control group. It has already supplied useful information about the genetic basis of asthma, cancer, diabetes, heart disease and mental illness. GWASs, however, assume that the two groups being compared are drawn from the same homogeneous population—that is, there is no underlying pattern of genetic difference between them other than susceptibility to the disease condition being tested.
Although the population as a whole does not show great genetic diversity, previous work by other researchers suggested that the Japanese fall into two genetic categories, perhaps reflecting two migration events. But these studies were of limited regions of the genome. So the RIKEN researchers embarked on a much broader investigation with greater relevance to GWASs. They published their results in a recent issue of The American Journal of Human Genetics1.
Given the good quality of SNP data on the Japanese population, the researchers used statistical techniques mainly based on principal components analysis to check the homogeneity of genetic structure in a sample of 7,003 people drawn from all over Japan. They found strong evidence of the dual nature of the Japanese population. One genetic group, the Hondo cluster, includes most people from the main islands of Japan; the other, the much smaller Ryukyu cluster, includes most individuals from Okinawa and nearby islands.
The research also demonstrated how such population stratification could impact the results of GWASs using Japanese subjects. As a consequence, the researchers propose measures to avoid bias, such as excluding members from the Ryukyu cluster if they occur in small proportion, equalizing Ryukyu numbers in both disease and control groups, or using statistical techniques to compensate for any potential bias.
“The aim of our analysis is to improve methods for conducting GWASs,” says first author Yumi Yamaguchi-Kabata. “We now wish to broaden the work to encompass different local regions in Asia.”
References
- 1. Yamaguchi-Kabata, Y., Nakazono, K., Takahashi, A., Saito, S., Hosono, N., Kubo, M., Nakamura, Y. & Kamatani, N. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: Effects on population-based association studies. The American Journal of Human Genetics 83, 445–456 (2008). doi: 10.1016/j.ajhg.2008.08.019
