Jan. 30, 2009 Research Highlight Biology
Staying true to the code
By turning enzymes into better editors, Japanese scientists achieve improved results in protein engineering
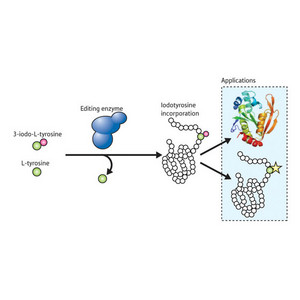 Figure 1: Schematic of the synthetase editing functionality. Synthetase enzymes with a clear preference for 3-iodo-L-tyrosine will occasionally introduce L-tyrosine by accident. The integration of an editing domain prevents this by cutting L-tyrosine molecules loose, so that proteins synthesized from RNAs containing the appropriate codon will consistently incorporate only the modified amino acid.
Figure 1: Schematic of the synthetase editing functionality. Synthetase enzymes with a clear preference for 3-iodo-L-tyrosine will occasionally introduce L-tyrosine by accident. The integration of an editing domain prevents this by cutting L-tyrosine molecules loose, so that proteins synthesized from RNAs containing the appropriate codon will consistently incorporate only the modified amino acid.
Cells achieve astonishing protein diversity with a fairly limited palette: just 20 naturally occurring amino acids. However, there is sufficient flexibility inherent in the protein-coding and -translation processes that has allowed enterprising scientists to develop clever means for introducing additional, unnatural amino acids into otherwise normal proteins.
As cellular translation machinery reads an RNA transcript, it identifies nucleotide triplets known as codons that indicate what amino acid is needed—these are then delivered through the assistance of a family of enzymes called aminoacyl-tRNA synthetases, which ensure proper placement of the proper molecule.
In previous work, Shigeyuki Yokoyama from the RIKEN Systems and Structural Biology Center in Yokohama and colleagues demonstrated that these enzymes can be engineered to introduce alternative amino acids into proteins1—in this case, substituting L-tyrosine with 3-iodo-L-tyrosine, a synthetic analog with uses in imaging and structural biology applications.
Unfortunately, this approach was not a total success, as the mutated synthetase enzyme (iodoTyrRS) still processes L-tyrosine at a reduced, but still significant, frequency. “Any further changes made in the amino-acid-binding pocket [of the enzyme] could not reduce the error rate,” explains Kensaku Sakamoto, a member of Yokoyama’s team. “We had to change the approach to achieve satisfactory selectivity.”
The team’s solution was to teach iodoTyrRS to be a better proof-reader. In new work, Yokoyama and colleagues have further engineered a version of the synthetase, one that incorporates a transplanted enzymatic domain that specifically recognizes and purges L-tyrosine2.
They began by analyzing various synthetase enzymes that exhibit this editing functionality, in an effort to zoom in on the domains that confer this ability. Once these had been identified, they grafted these domains onto different sections of iodoTyrRS. One of the variants, CP-Ped-IYRS, showed strong specificity for 3-iodo-L-tyrosine alone (Fig. 1), while the original iodoTyrRS enzyme and other variants would regularly make use of L-tyrosine when the synthetic amino acid was no longer available. This specificity was greatly reduced when the editing domain was inactivated by mutation, indicating that this functionality is highly important for establishing strict amino acid preference.
The team is now looking to move their modified enzymes out of the test tube and into living cells, but they are generally quite pleased with this demonstration of grafting two sophisticated enzymatic activities together seamlessly. “Our approach may facilitate developing other enzymes required for incorporating non-natural amino acids into proteins site-specifically,” says Sakamoto, “so that the list of available non-natural amino acids can thus be further extended.”
References
- 1. Kiga, D., Sakamoto, K., Kodama, K., Kigawa, T., Matsuda, T., Yabuki, T., Shirouzu, M., Harada, Y., Nakayama, H., Takio, K. et al. An engineered Escherichia coli tyrosyl–tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proceedings of the National Academy of Sciences USA 99, 9715–9720 (2002). doi: 10.1073/pnas.142220099
- 2. Oki, K., Sakamoto, K., Kobayashi, T., Sasaki, H. & Yokoyama, S. Transplantation of a tyrosine editing domain into a tyrosyl-tRNA synthetase variant enhances its specificity for a tyrosine analog. Proceedings of the National Academy of Sciences USA 105, 13298–13303 (2008). 10.1073/pnas.0803531105
