Nov. 6, 2009 Research Highlight Biology
Getting membrane proteins into the fold
A new preparation method promises to bring a challenging but clinically important subset of proteins within easier reach of scientists
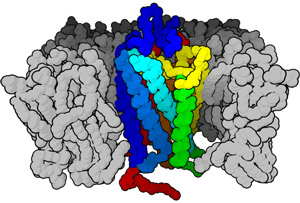 Figure 1: Schematic of a membrane-bound protein, which relies on the hydrophobic environment within the cell membrane. These proteins are notoriously difficult to overexpress and purify in large quantities. Source: Wikipedia
Figure 1: Schematic of a membrane-bound protein, which relies on the hydrophobic environment within the cell membrane. These proteins are notoriously difficult to overexpress and purify in large quantities. Source: Wikipedia
Membrane-embedded proteins execute many of the most important cellular functions, detecting external environmental cues, mediating communication and signaling, and facilitating molecular import and export. Accordingly, they also represent important targets for drug development, and an estimated 60% of currently available drugs are believed to target membrane proteins.
On the other hand, experimental investigation of the structure and function of individual membrane proteins is routinely thwarted by the general difficulty of preparing large quantities of properly folded protein. This is largely due to the chemical composition of these proteins, which have large hydrophobic surfaces that are chemically averse to being exposed to water; these protein segments are stable when embedded within the equally hydrophobic lipid-based cell membrane (Fig. 1), but tend to assemble into irregular, improperly folded clumps when prepared in solution.
Most water-soluble proteins can readily be produced either within modified cell lines or through the use of cellular extracts that contain the full complement of machinery required for protein synthesis. However, the yield of membrane proteins obtained from cultured cells is generally inadequate, and scientists have had to tinker extensively with extract-based production methods to obtain usable quantities of functional protein.
Now, a research team led by Shigeyuki Yokoyama at the RIKEN Systems and Structural Biology Center in Yokoyama has developed an efficient preparation method that produces high yields of functional membrane proteins1. “Up until now, aggregation and misfolding of membrane proteins during cell-free protein synthesis have been avoided by adding either lipid-based liposomes or detergents,” explains Yokoyama. However, he adds that such workarounds were not a complete solution, and challenges remained in selecting the appropriate detergent for a given protein, or ensuring that newly synthesized proteins were properly folded and inserted into the membranes of liposomes.
Best of both worlds
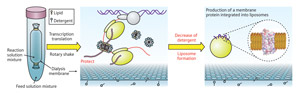 Figure 2: An overview of the new protein production process. Protein is produced in a tube via a cell-free transcription and translation reaction (left). This reaction is performed in the presence of lipids and detergent, which protect hydrophobic protein segments and help initial folding along (middle). When detergent is removed by dialysis, the properly folded proteins end up embedded within liposomes, replicating their normal integration into cell membranes (right). © (2009) RIKEN
Figure 2: An overview of the new protein production process. Protein is produced in a tube via a cell-free transcription and translation reaction (left). This reaction is performed in the presence of lipids and detergent, which protect hydrophobic protein segments and help initial folding along (middle). When detergent is removed by dialysis, the properly folded proteins end up embedded within liposomes, replicating their normal integration into cell membranes (right). © (2009) RIKEN
Yokoyama and colleagues solved this problem by developing a hybrid technique that incorporates elements of both preparation methods, synthesizing their proteins in a bacteria-derived extract containing both detergent and lipid molecules. During production, hydrophobic stretches of the newly formed protein are protected by this lipid-detergent mixture; once synthesis is complete, the detergent is removed from the sample via dialysis and the remaining lipids subsequently assemble into bubble-shaped liposomes with the mature proteins securely embedded within these artificial membranes (Fig. 2).
The researchers then demonstrated the efficacy of their technique with bacteriorhodopsin (BR), an archaea-derived photosynthetic pigment protein. The light-responsive properties of BR made it a particularly useful test subject for this method—improperly folded or aggregated BR appears yellow, while preparations of mature BR appear purple. In initial experiments, they were able to demonstrate successful production of properly folded BR with a variety of detergents, and although the overall folding efficiency was somewhat lower than previously described preparations, the overall quantity of protein produced was significantly greater—as much as 80-fold greater, depending on the detergent. Subsequent analysis confirmed that much of this properly folded BR was successfully integrated into the membranes of liposomes following detergent removal.
Yokoyama and his colleagues' protein preparations also retained proper functional characteristics. Naturally occurring BR undergoes a series of chemical transitions known as a photocycle, in which excitation by light at particular wavelengths induces a series of subtle structural changes; these shifts can in turn be quantified by changes in the light-absorption properties of the protein. They found that their synthetic BR mirrored the photocyclic characteristics of native protein, and even displayed the capacity to act as a light-activated proton pump—a key component of its photosynthetic activity.
Making a big production of it
Whether researchers are looking to derive protein in crystalline form for structural analysis or in solution for functional characterization, the ability to obtain high-purity preparations is essential. In this regard, the Yokoyama team’s technique passed with flying colors, with a 75-fold greater yield of functional BR protein relative to a previously described high-yield liposome-based method.
Yokoyama indicates that since the initial publication of this work, his group has had the opportunity to test their method on a variety of membrane-bound receptors and channels, and they are currently in the process of characterizing the efficiency of these preparations. Fortunately, the system is sufficiently flexible that a number of adaptations can be introduced to optimize production for any given protein. “We will test other lipids or steroid detergents for the functional overproduction of other types of membrane proteins,” he says. “Moreover, by controlling lipid bilayer formation and protein synthesis speed through changes in reaction conditions such as temperature, we can improve this system to be suitable for many types of membrane proteins.”
Even as Yokoyama and his colleagues work out the details of refining their overexpression system, they have already begun to contemplate future targets for which this method might prove valuable—and with so many poorly characterized membrane proteins, the field is wide open. “We hope to apply this system to biochemical, biophysical and structural studies of human G-protein coupled receptors, which are important targets for drug design, and proteins that transport small molecules such as nutrients, across the membrane,” he says, “as well as complexes incorporating membrane proteins, because the cell-free system makes it easy to co-express more than one protein.”
References
- 1. Shimono, K., Goto, M., Kikukawa, T., Miyauchi, S., Shirouzu, M., Kamo, N. & Yokoyama, S. Production of functional bacteriorhodopsin by an Escherichia coli cell-free protein synthesis system supplemented with steroid detergent and lipid. Protein Science 18, 2160–2171 (2009). doi: 10.1002/pro.230
About the Researcher
Shigeyuki Yokoyama
Shigeyuki Yokoyama was born in Tokyo, Japan, in 1953. He received his BS and PhD degrees from The University of Tokyo in 1975 and 1981, and following completion of five years of postdoctoral work, became an associate professor in 1986 and a professor in 1991 in the Department of Biophysics and Biochemistry, The University of Tokyo. In 1993, he was appointed chief scientist of the RIKEN Cellular Signaling Laboratory, and later project director of the Protein Research Group in the Genomic Sciences Center. He played a pivotal role as science director of the RIKEN Structural Genomics / Proteomics Initiative (RSGI). Since 2008, he has acted as director of the Systems and Structural Biology Center (SSBC) at the RIKEN Yokohama Institute.
