Dec. 25, 2009 Research Highlight Biology
Getting past inspection
By being picky about its binding partners, an RNA-modifying enzyme brings an important measure of quality control to the protein production process
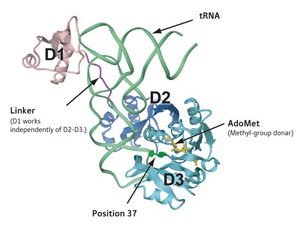 Figure 1: Schematic of the structure of the aTrm5 complex showing that the tRNA residue G37 (bright green) is positioned near the methyl-donor AdoMet within the catalytic domain (D2 and D3 of the enzyme), while sensor domain D1 recognizes the tRNA ‘corner’ of the L-shaped structure. © (2009) RIKEN
Figure 1: Schematic of the structure of the aTrm5 complex showing that the tRNA residue G37 (bright green) is positioned near the methyl-donor AdoMet within the catalytic domain (D2 and D3 of the enzyme), while sensor domain D1 recognizes the tRNA ‘corner’ of the L-shaped structure. © (2009) RIKEN
Messenger RNA (mRNA) molecules are the key intermediaries that enable translation of gene-encoded information into functional protein, but this process also relies on another essential family of RNAs—the various transfer RNAs (tRNAs) that physically ‘read’ the mRNA sequence and facilitate delivery and addition of appropriate amino acids to the nascent protein chain.
Although there is a specific tRNA for each individual amino acid, they all share a common L-shaped structure, which has a so-called anticodon mRNA-reading domain and an amino-acid-laden acceptor domain located at opposite ends. The assembly of a mature, functional tRNA depends on the addition of numerous targeted chemical modifications by a variety of specialized enzymes, such as Trm5, which tacks a methyl chemical group onto a specific guanine nucleotide in the anticodon region. For many tRNAs, this Trm5 modification is a crucial step, and new research from Shigeyuki Yokoyama and colleagues at the RIKEN Systems and Structural Biology Center in Yokohama has revealed unexpected insights into why this is the case1.
The researchers began by collecting high-resolution structural data about the complex formed by Trm5 from the archaea Methanocaldococcus jannaschii (aTrm5) with a target tRNA and the methyl-group donor molecule AdoMet. As expected, their analysis revealed that the target guanine of the tRNA, G37, is positioned at a highly exposed orientation near the AdoMet molecule’s methyl group at the catalytic center of aTrm5.
However, Yokoyama’s team also characterized additional interactions between an enzymatic ‘sensor’ domain and structural elements located at the corner of the tRNA ‘L’, suggesting that aTrm5 selectively interacts only with fully folded target molecules (Fig. 1). Follow-up biochemical analyses confirmed the importance of this interaction in facilitating catalysis; at temperatures between 70 and 80 °C, where M. jannaschii typically thrives, aTrm5 variants entirely lacking the sensor domain exhibited dramatically reduced tRNA methylation activity relative to full-length enzyme, as did aTrm5 mutants with mutations at specific corner-binding positions.
Given the fundamental importance of G37 methylation in preventing ‘typos’ during protein production and the strong structure-dependence of Trm5 tRNA recognition and catalysis, Yokoyama and colleagues hypothesize that this enzyme may play a vital role in quality control. “The tRNA maturation process has a ‘checkpoint’ composed of the aTrm5 sensor-effector system to verify whether the tRNA assumes the proper L shape, in order to send only qualified tRNAs out to the translational stage,” they conclude, adding that this may ultimately represent just one of many such ‘inspection’ steps over the course of maturation.
References
- 1. Goto-Ito, S., Ito, T., Kuratani, M., Bessho, Y. & Yokoyama, S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nature Structural & Molecular Biology 16, 1109–1115 (2009). doi: 10.1038/nsmb.1653
