Mar. 12, 2010 Research Highlight Biology
All natural ingredients
A catalog of the chemicals produced within a plant’s tissues yields fresh insights into its metabolic pathways and gene function
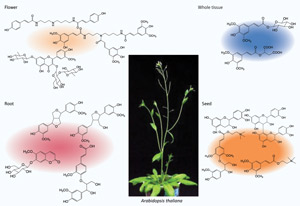 Figure 1: As revealed by the AtMetExpress project, various tissues of Arabidopsis thaliana contain compounds associated with metabolism that are diverse in structure. © 2010 Fumio Matsuda
Figure 1: As revealed by the AtMetExpress project, various tissues of Arabidopsis thaliana contain compounds associated with metabolism that are diverse in structure. © 2010 Fumio Matsuda
The various metabolic pathways in a given plant generate a staggering array of molecules that enable growth and survival under diverse conditions—and in some cases, hold value for scientific applications ranging from pharmaceutical research to the development of new materials. “The huge chemical diversity of plants exceeds that of most animals and microorganisms,” says Kazuki Saito of the RIKEN Plant Science Center in Yokohama.
The thale cress, Arabidopsis thaliana, is a widely used model for genetic and developmental research and possibly the best characterized of all plant species; nevertheless, scientists remain far from completing a comprehensive ‘metabolome’, or atlas of metabolites, for this organism. The AtMetExpress project, launched by Saito and colleagues, seeks to rectify this situation by assembling a massive, annotated roster of molecules gathered from 36 different Arabidopsis tissue samples1.
“What we needed was the pattern of metabolite accumulation during plant development to understand cell function more precisely,” explains Saito. To start with, his team used a method called liquid chromatography-mass spectrometry (LC-MS) to derive information about the chemical content of a variety of plant organs collected at different developmental stages. The researchers subsequently cross-referenced these against a library of tandem mass spectrometry spectral tags (MS2Ts)—essentially an index of the individual compounds that can be detected in Arabidopsis. By this process, they were able to assign unique MS2Ts to approximately 95% of the molecules detected via LC-MS and subsequently managed to derive structural information for a total of 167 metabolites (Fig. 1).
A comparison of the spatial and temporal distributions for these various metabolites with detailed datasets describing gene expression in Arabidopsis allowed Saito and colleagues to obtain new insights into metabolic regulation, revealing several especially complex pathways where levels of a given metabolite were seemingly decoupled from expression levels of the genes involved in its synthesis, suggesting the existence of potentially diverse additional modes of control.
Saito and colleagues were also able to assign roles to previously unknown putative components of biosynthetic pathways. “We found a number of correlations between metabolite peaks and uncharacterized genes,” says Saito. “These are potential targets for discovery of new genes involved in metabolite production.”
This first iteration of AtMetExpress will serve as a foundation for more targeted future investigations, including the exploration of metabolic pathways triggered by the activity of specific plant hormones, and ultimately may lead the way for similar analyses of other plant species with important agricultural or medicinal applications.
References
- 1. Matsuda, F., Hirai, M.Y., Sasaki, E., Akiyama, K., Yonekura-Sakakibara, K., Provart, N.J., Sakurai, T., Shimada, Y. & Saito, K. AtMetExpress development: A phytochemical atlas of Arabidopsis thaliana development. Plant Physiology 152, 566–578 (2010). doi: 10.1104/pp.109.148031
