Jul. 16, 2010 Research Highlight Biology
Mapping brain development
A large-scale genetic analysis provides a molecular atlas of a complex brain structure, the hypothalamus
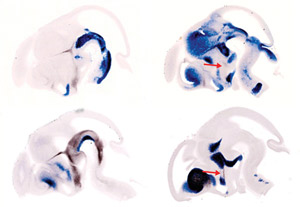 Figure 1: Light microscope images of four different embryos, showing longitudinal whole brain sections viewed from the side, at the same developmental stage. Molecular markers delineate the borders of distinct nuclei, or groups of neurons, in the hypothalamus (bottom center of each brain section). Sonic Hedgehog (dark brown) in the left column is required for differentiation of cells in the nuclei at the front of the hypothalamus. The function of another marker, Lhx6 (dark brown, red arrows) in the right column is in now under investigation. © 2010 Tomomi Shimogori
Figure 1: Light microscope images of four different embryos, showing longitudinal whole brain sections viewed from the side, at the same developmental stage. Molecular markers delineate the borders of distinct nuclei, or groups of neurons, in the hypothalamus (bottom center of each brain section). Sonic Hedgehog (dark brown) in the left column is required for differentiation of cells in the nuclei at the front of the hypothalamus. The function of another marker, Lhx6 (dark brown, red arrows) in the right column is in now under investigation. © 2010 Tomomi Shimogori
The hypothalamus is a small part of the brain that regulates many behaviors that are critical for survival, including circadian rhythms, food intake, temperature regulation and the release of hormones from the pituitary gland. Abnormal development of this structure causes hormonal imbalances thought to underlie multiple diseases, including autism, mood disorders and obesity.
Anatomically, the hypothalamus is highly complex, with many distinct nuclei each containing a cluster of unique cells. Since little is known about the molecular mechanisms mediating the development of this well-characterized structure, Tomomi Shimogori of the RIKEN Brain Science Institute, Wako, and her multinational colleagues performed a genome-wide DNA microarray analysis to investigate gene expression at 12 different intervals during development of the mouse hypothalamus1.
They found that, between embryonic days 10 and 16 when immature neurons are being generated by cell division, the expression of 1,045 genes is enhanced. The researchers also noted that expression of some of these genes, which are known to be expressed in all immature neurons, peaked early then declined steadily. Those expressed uniquely in the hypothalamus peaked between embryonic days 12 and 14, when the production of hypothalamic cells also peaks.
After examining the expression patterns of all 1,045 genes, the researchers used the data to produce a molecular atlas, showing when and where each is expressed in the hypothalamus. This not only revealed several molecular ‘markers’ that label each of the hypothalamic nuclei throughout the entire course of development, but also delineated their borders (Fig. 1).
One gene, Sonic Hedgehog (Shh), is well known to be expressed along the bottom of the developing neural tube and to induce a specific pattern of motor neurons in that region. Shimogori and her colleagues found another Shh expression domain, at the base of the developing hypothalamus, and speculated that it might also be involved in cell patterning.
To investigate, they created genetically engineered mice in which Shh is selectively deleted from the hypothalamus. The hypothalamus was notably thinner in these animals, and expression of several genes, normally found later on in nuclei at the front of the hypothalamus, was missing. This suggests that cells at the front of the hypothalamus fail to differentiate in the absence of Shh.
“The next step is identifying the genes responsible for shaping the hypothalamus,” says Shimogori. “To do this, we are currently overexpressing patterning molecules, analyzing the effects and using our gene atlas to predict their role.”
References
- 1. Shimogori, T., Lee, D. A., Miranda-Angulo, A., Yang, Y., Wang, H., Jiang, L., Yoshida, A. C., Kataoka, A., Mashiko, H., Avetisyan, M., Qi, L., Qian, J. & Blackshaw, S. A genomic atlas of mouse hypothalamic development. Nature Neuroscience 13, 767–775 (2010). doi: 10.1038/nn.2545
