Oct. 16, 2015 Research Highlight Biology
Knotty problem solved
The knotted structure of an enzyme that modifies transfer RNAs has been untied
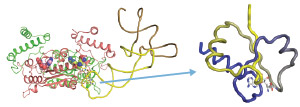 Figure 1: Left, ternary complex consisting of bacterial transfer RNA (tRNA) methyltransferase D (TrmD), a methyl group donor and tRNA. Right, the knot of the TrmD folds the methyl donor rigidly. Reproduced, with permission, from Ref. 1 © 2015 T. Ito et al.
Figure 1: Left, ternary complex consisting of bacterial transfer RNA (tRNA) methyltransferase D (TrmD), a methyl group donor and tRNA. Right, the knot of the TrmD folds the methyl donor rigidly. Reproduced, with permission, from Ref. 1 © 2015 T. Ito et al.
RIKEN researchers have deciphered the knotted structure of an enzyme that assists the translation of genetic code into proteins1. It had been believed that proteins do not form knots, until Shigeyuki Yokoyama and colleagues identified a family of enzymes known as RNA methyltransferases, which help translate genetic information into proteins. These enzymes form a deep trefoil knot, a loop in which the two loose ends of a simple knot are tied together through tertiary interactions.
The researchers wanted to uncover the fundamental mechanisms of protein translation; in particular, how the trefoil knot structure of the bacterial transfer RNA (tRNA) methyltransferase D (TrmD) contributes to this process.
TrmD forms a ternary complex with a methyl group donor and the tRNA (Fig. 1) and then transfers the methyl group to the guanosine in position 37 (G37) on the tRNA. Each tRNA carries an amino acid that corresponds in the genetic code to the three bases in its anticodon; these bases bind the codon, the complementary three bases on the messenger RNA (mRNA). The ribosome catalyzes the addition of the amino acid to the growing peptide chain and a protein is formed.
Methylation at G37 ensures that the tRNA correctly binds to the codon, so that the genetic information is translated faithfully. Loss of G37 methylation can lead to ‘frameshifting’, so-called because the tRNA slides one base past the intended codon on the mRNA, thereby shifting the reading frame and often producing nonfunctional proteins.
“Methyl-G37 modification is one of the best-studied tRNA modifications because of its biological importance,” explains Yokoyama, who is at the RIKEN Structural Biology Laboratory. “Therefore, the tRNA-bound structure of TrmD has been awaited for many years.”
In collaboration with scientists in the United States and South Korea, his RIKEN team solved the crystal structure of ternary complexes containing bacterial TrmD, a methyl donor and a tRNA. They discovered how the enzyme’s structure changes during the enzymatic reaction: TrmD captures the methyl donor in its loose-knot stage and then strengthens its grip on it by tightening the knot. The rigidly fixed methyl donor then causes large structural changes in the protein that activate both the tRNA-binding and catalytic sites, and the methyl group is transferred to G37 of the bound tRNA.
“We showed that the trefoil knot is essential for the sophisticated mechanisms underlying the enzyme function,” says Yokoyama.
Yokoyama’s team is now investigating how the lack of G37 methylation affects global protein synthesis in bacteria, research that may lead to the development of potent, side-effect-free antimicrobial agents.
References
- 1. Ito, T., Masuda, I., Yoshida, K.-i., Goto-Ito, S., Sekine, S.-i., Suh, S. W., Hou, Y.-M. & Yokoyama, S. Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proceedings of the National Academy of Sciences USA 112, E4197–E4205 (2015) doi: 10.1073/pnas.1422981112
