Jun. 6, 2014 Press Release Biology
Cutting edge methods reveal what makes Purkinje neurons unique
In a collaboration between RIKEN's Brain Science Institute and Center for Life Science Technologies in Japan, scientists combined cutting edge methods to obtain a comprehensive catalog of proteins that are manufactured in specific parts of Purkinje neurons.
The study, headed by Drs. Thomas Launey, Unit Leader at RIKEN BSI and Charles Plessy, Unit Leader at RIKEN CLST, and published in Genome Research, succeeded in identifying several thousand RNAs that are enriched in rat Purkinje neurons. This comprehensive list holds the key to a better understanding of molecular events within these neurons and potential insights into diseases associated with Purkinje cells, like ataxias (disorders affecting movement) and autism.
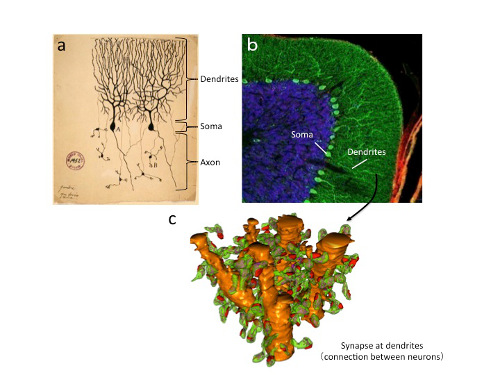
"Purkinje neurons, remain one of most mysterious type of brain cell, receiving 10 times more connections than any other neuron and possessing structural characteristics that are unique among neurons," said Dr. Launey. These large neurons have a distinctive shape with extensively branched extensions called dendrites, which receive signals, and a single axon for sending signals to other neurons. Unlike other inhibitory neurons that act locally on their neighbors, the Purkinje cells shut off neurons that are located outside of the cerebellar cortex.
Drs. Anton Kratz and Pascal Beguin, the study's co-first authors, and colleagues identified not only what proteins distinguish Purkinje cells from other neurons, but also what proteins are made directly in the dendrites. Isolating the dendrites and examining their contents is challenging, and the researchers accomplished this technical feat using sophisticated methods.
Although all cells of the body have the same DNA, they make different proteins, which give them unique properties, depending on what parts of the genome are active. To make the proteins, messenger RNA (mRNA) carries information encoded in genes from the nucleus to the ribosomes, the protein-building machinery in the cytoplasm. By combining two high-throughput methods known as TRAP (Translating Ribosomal Affinity Purification) and CAGEscan (Cap Analysis of Gene Expression, developed at RIKEN), researchers could specifically capture and identify the mRNAs as they were being translated into proteins by ribosomes, in rat Purkinje neurons.
The TRAP method introduces a tag into the ribosome and the researchers used biochemical methods to purify the tagged ribosomes with the bound mRNA specifically from Purkinje cells, revealing what proteins are being manufactured. CAGEscan then allowed them to quickly sequence and identify the huge number of trapped mRNAs. Using microdissection to separate the cell body from the tree-like dendrites allowed the researchers to identify the proteins unique to each compartment. They found that there are many different kinds of proteins manufactured specifically in the dendrites that were distinct from the cell body.
This near-exhaustive survey of the molecular landscape allowed the researchers to find novel identifiers of Purkinje neurons and previously unknown mRNAs. "The introduction of new methods gave us access to mRNAs that are often difficult to detect, especially those that are in low abundance, but are functionally important, such as membrane receptors, transporters and ion channels," said Dr. Launey.
The methods used in this study should be widely applicable to other neuron types and other mammals, including primates, paving the way for comparative studies that may reveal the origins of complex cognitive abilities or the causal defects in diseases affecting the brain.
Reference
- Anton Kratz, Pascal Beguin, Megumi Kaneko, Takahiko Chimura, Ana Maria Suzuki, Atsuko Matsunaga, Sachi Kato, Nicolas Bertin, Timo Lassmann, Rejan Vigot, Piero Carninci, Charles Plessy and Thomas Launey. "Digital expression profiling of the compartmentalized translatome of Purkinje neurons". Genome Research, 2014, doi:10.1101/gr.164095.113
Contact
Unit Leader
Thomas Launey
Launey Resarch Unit
RIKEN Brain Science Institute
Unit Leader
Charles Plessy
Genomics Miniaturization Technology Unit
Life Science Accelerator Technology Group
Division of Genomic Technologies
RIKEN Center for Life Science Technologies
Jens Wilkinson
RIKEN Global Relations and Research Coordination Office
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Email: pr@riken.jp
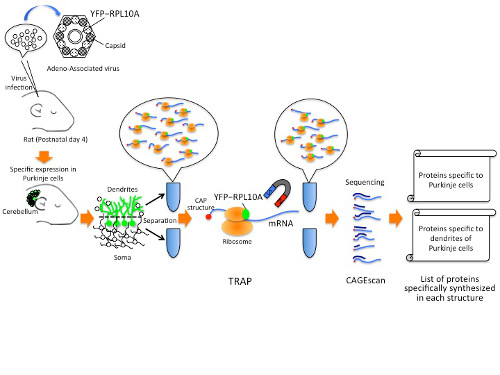
Schematic of the methods used to isolate proteins from Purkinje cells and their dendrites
