Jan. 12, 2015 Press Release Biology
Revealing the inner workings of a molecular motor
In research published in the Journal of Cell Biology, scientists from the RIKEN Brain Science Institute in Japan have made important steps toward understanding how dynein—a "molecular motor"—walks along tube-like structures in the cell to move cellular cargo from the outer structures toward the cell body of neurons. The action of this molecule is important for a number of cell functions including axonal transport and chromosome segregation, and its dysfunction is known to lead to a congenital developmental brain disorder known as lissencephaly.
Though cells may look like shapeless blobs of liquid encased in a membrane, in fact they have a complex skeleton-like structure, known as the cytoskeleton, made up of filaments called microtubules. Motor proteins, which include dynein and kinesin, can move along these tubules to transport cargo into and out of the center of the cell. The motor proteins use an energy-currency molecule, ATP, to power their movements along the microtubules. The motor proteins hydrolyze ATP to ADP, and convert the released chemical energy to mechanical energy which is used for movement. The mechanism is quite well understood for kinesin, but in the case of dynein, it has been difficult to explain how communication takes place between the site of microtubule binding and the site of ATP hydrolysis, which are relatively far from each other, separated by a stalk.
In the new research, performed in collaboration with several other institutes including the University of Osaka, Waseda, and Hosei University, the RIKEN scientists used cryo electron microscopy—where molecules are cooled to very low temperatures in the microscope—and examined the structure of dynein on the microtubule. They showed that two specific amino acid residues on the microtubule structure, R403 and E416, are key to turning on the switch that is critical for the activation of the dynein motor—demonstrating that when mutations in these sequences are present, the dynein fails to achieve directional movement on the microtubule, ending up simply moving back and forth in a random fashion. This lends weight to the idea, that has been generally accepted, that the motion of molecular motors is basically driven by random, Brownian motion, and that motors are able to move in one direction thanks to subtle changes in the strength of bonds at the motor-microtubule interface.
Additionally, the group discovered that turning on the mechanical switch at the motor-microtubule interface leads to ATP hydrolysis. Their results altogether indicate that the subtle structural changes in the bonds at the interface are transmitted through a small change in the structure of the stalk—there are two coils that link the two binding regions, and a small shift in the configuration of the coils gives the cue for ATP hydrolysis at the ATP binding site.
Seiichi Uchimura, the first author of the paper, said, "We were able to clearly demonstrate that the dynein molecular motor is activated by a 'switch' that controls mutual interactions between dynein and the microtubule. This is important, as a mutation in the structure of the switch has been demonstrated to cause lissencephaly, a congenital disorder."
According to Etsuko Muto, who led the research team, "In the future, we hope that further understanding the interplay between dynein and microtubule, as this could pave the way for therapies for these conditions."
Reference
- Uchimura, S. Fujii, T. Takazaki, H. Ayukawa, R. Nishikawa, Y. Minoura, I. Hachikubo, Y. Kurisu, G. Sutoh, K. Kon, T. Namba, K. and Muto, E., "A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation", The Journal of Cell Biology, doi: 10.1083/jcb.201407039
Contact
Etsuko Muto, Laboratory Head
Seiichi Uchimura
Laboratory for Molecular Biophysics
RIKEN Brain Science Institute
Jens Wilkinson
RIKEN Global Relations and Research Coordination Office
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Email: pr@riken.jp
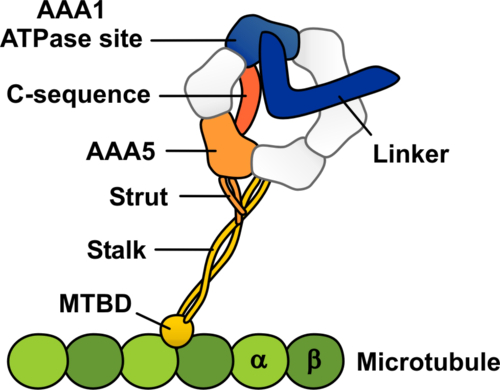
Schematic representation of the dynein-microtubule complex showing the structural elements likely to be involved in allosteric communication between the microtubule and the ATPase site in dynein
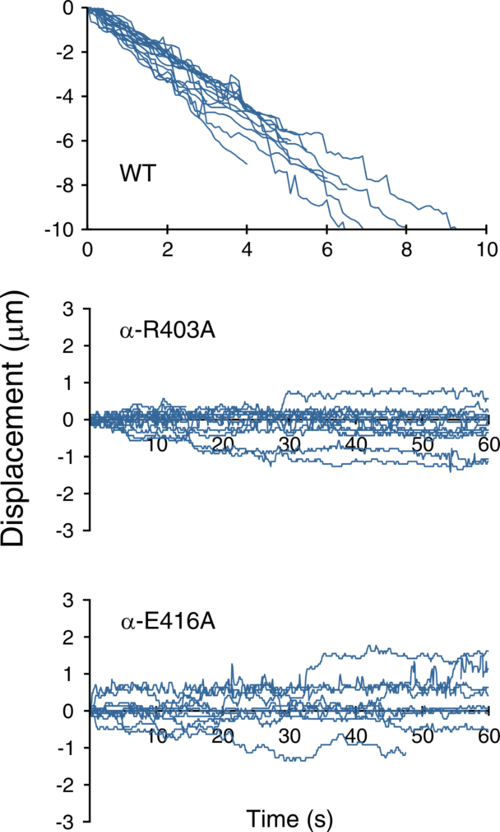
Graph showing the movement over time of wild type and mutant types along the microtubules in the presence of 1 mM ATP. The mutants can be seen to be going back and forth.
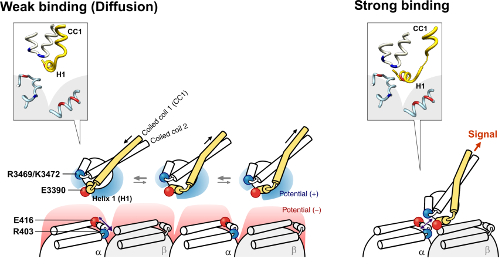
The figures on the left and right show a model of the microtubule binding domain-microtubule interaction in the weak and strong binding states, respectively.
