Mar. 2, 2017 Press Release Biology
Improved gene expression atlas shows that many human long non-coding RNAs may actually be functional
While it was once believed that genes regulated biological functions almost exclusively by being transcribed to coding RNAs that were then translated into proteins, it is now known that the picture is much more complex. In fact, studies examining the association between genes and diseases have shown that most disease variants are found outside of protein-coding genes.
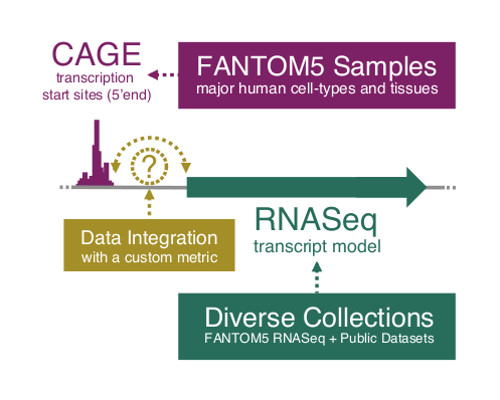
The RIKEN-led FANTOM consortium pioneered the discovery of non-coding RNAs over a decade ago, revealing the complexity of the transcriptional landscape in mammalian genomes for the first time. The FANTOM consortium continues to be on the leading edge of studies into the origins and functions of non-coding RNAs. In their latest work, published in Nature, the team has generated a comprehensive atlas of human long non-coding RNAs with substantially improved gene models, allowing them to better assess the diversity and functionality of these RNAs. Most attempts today to draw maps of RNA transcription rely on sequencing technologies that do not always accurately identify the beginnings, or 5’ ends, of the RNA transcripts. To overcome this limitation, the team used a technology known as Cap Analysis of Gene Expression (CAGE), which was developed at RIKEN, to build an atlas of human long non-coding RNAs with accurate 5’ ends, precisely pinpointing where in the genome their transcription is initiated.
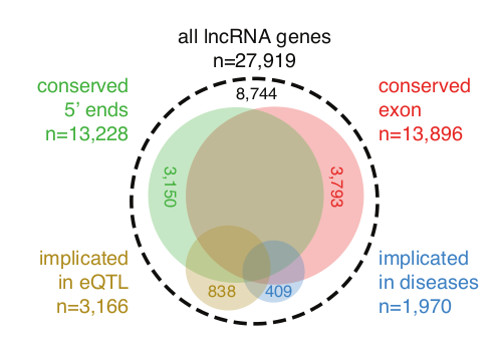
The atlas, which contains 27,919 long non-coding RNAs, summarizes for the first time their expression patterns across the major human cell types and tissues. By intersecting this atlas with genomic and genetic data, their results suggest that 19,175 of these RNAs may be functional, hinting that there could be as many—or even more—functional non-coding RNAs than the approximately 20,000 protein-coding genes in the human genome.
“There is strong debate in the scientific community on whether the thousands of long non-coding RNAs generated from our genomes are functional or simply byproducts of a noisy transcriptional machinery.” says Professor Alistair Forrest of the Harry Perkins Institute of Medical Research at the University of Western Australia and Senior Visiting Scientist at the RIKEN Center for Life Science Technologies (CLST), one of the corresponding authors of the paper, “By integrating the improved gene models with data from gene expression, evolutionary conservation and genetic studies, we find compelling evidence that the majority of these long non-coding RNAs appear to be functional, and for nearly 2,000 of them we reveal their potential involvement in diseases and other genetic traits.” “Intriguingly,” says Chung-Chau Hon of CLST, first author on the paper, “the majority of long non-coding RNAs appear to be generated from enhancer elements. It deepens our understanding towards the largely heterogeneous origins of long non-coding RNAs.”
According to Piero Carninci of CLST, “The improved gene models and the broad functional hints of human long non-coding RNAs derived from this atlas could serve as a Rosetta Stone for us to experimentally investigate their functional relevance as part of our ongoing work for the upcoming edition of the FANTOM consortium. We anticipate that these results could further push the boundary of our understanding of the functions of the non-coding portion of our genome.”
The resources of the long non-coding RNA atlas are available at FANTOM website.Reference
- Chung-Chau Hon, Jordan A. Ramilowski, Jayson Harshbarger, Nicolas Bertin, Owen J L Rackham, Julian Gough, Elena Denisenko, Sebastian Schmeier, Thomas M. Poulsen, Jessica Severin, Marina Lizio, Hideya Kawaji, Takeya Kasukawa, Masayoshi Itoh, A. Maxwell Burroughs, Shohei Noma, Sarah Djebali, Tanvir Alam, Yulia A. Medvedeva, Alison C Testa, Leonard Lipovich, Chi-Wai Yip, Imad Abugessaisa, Micka.l Mendez, Akira Hasegawa, Dave Tang, Timo Lassmann, Peter Heutink, Magda Babina, Christine A. Wells, Soichi Kojima, Yukio Nakamura, Harukazu Suzuki, Carsten O. Daub, Michiel J.L. de Hoon, Erik Arner, Yoshihide Hayashizaki, Piero Carninci, Alistair R R Forrest, "An atlas of human long non-coding RNAs with accurate 5’ ends", Nature, doi: 10.1038/nature21374
Contact
Team Leader
Piero Carninci
Genome Information Analysis Team
Life Science Accelerator Technology Group
Division of Genomic Technologies
RIKEN Center for Life Science Technologies
Program Director
Yoshihide Hayashizaki
RIKEN Preventive Medicine & Diagnosis Innovation Program
Jens Wilkinson
RIKEN International Affairs Division
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Email: pr[at]riken.jp