Apr. 18, 2017 Press Release Biology Chemistry
Atomic structure reveals how cells translate environmental signals
Culminating a nearly 10 year effort, researchers have determined the atomic resolution structure of a key molecule that translates signals from a cell’s local environment into a language that the cell can understand and use. The determination of the architecture of the Inositol Tris-Phosphate Receptor (IP3R) had long been considered a major goal in biomedical research because of its strategic role inside cells as a molecular train station for transferring signals that control many cell functions. The structure is expected to contribute to the development of better therapeutic approaches for many diseases. The work was conducted by a team at RIKEN Brain Science Institute under the direction of Professor Katsuhiko Mikoshiba, whose laboratory cloned the first IP3R gene in 1989.
In all living cells, chemical signals are harnessed for intracellular communication. The inositol 1,4,5-trisphosphate (IP3) is one such signal that binds to the IP3 receptor (IP3R) to release calcium ions (Ca2+) from intracellular Ca2+ stores such as the endoplasmic reticulum. The IP3R-embedded Ca2+ stores are distributed in various microdomains within cells and have pivotal roles in processes as diverse as neural communication, differentiation, plasticity, and metabolism. Of the three genes identified, the brain dominant type 1 IP3R (IP3R1) is genetically causative for spinocerebellar ataxia 15/16/29 and Gillespie syndrome, and regulates cellular waste disposal processes implicated in the etiology of neurodegenerative diseases including Alzheimer’s disease. Although the important roles of IP3R in normal and disease conditions are well known, understanding how IP3 signals trigger the opening of the Ca2+ channel was elusive.
The new IP3R1 crystal structure reveals a rich cosmos of atomic scale details on its function. IP3R1 is a micromachine of 20 nm in diameter that contains two functional sub-structures, an IP3 binding site and a Ca2+ channel pore. The distance from the IP3-binding site to the channel pore is 7 nm, the longest among similar ion channels, and the fundamental question of how IP3-binding physically opens the channel from a long range has been unanswered in the decades since the gene was cloned. X-ray crystallography of the large cytosolic domain of a mouse IP3R1 in the absence and presence of IP3, at the RIKEN SPring-8 ion beam factory, pinpointed a long-range mechanism involving an IP3-dependent global movement of a part of the receptor called the curvature α-helical domain that serves as a bridge between the cytosolic and channel domains. Mutagenesis of this bridge revealed the essential role of a leaflet structure in the α-helical domain that relays IP3 signals to the channel, and may help to explain how long-range coupling from IP3 binding to the Ca2+ channel occurs.
The findings reveal similarities and differences with a recently published report on the IP3R using a completely different method called cryo-electron microscopy. In the related study, a group led by Irina Serysheva from the University of Texas Health Science Center at Houston proposed that channel activation by IP3 may occur by direct binding of the C-terminus and IP3-binding domain and coupling from the IP3-binding domain to neighboring subunits. The current data disagree with these conclusions, instead suggesting that IP3-binding site to the leaflet region underlies the dynamic structural changes by IP3. A comparison of the two structures reveals agreement on an immobile part of the curvature helical domain and a variable arrangement of other helical domains. The authors hypothesize that the immobile section would act as a rigid-body conducting a torque from IP3-binding sites to the channel domain, whereas the flexible regions would contribute to the dynamic properties of IP3R function.
Resolving the long-standing mystery of long-range communication that allows IP3 to open the channel will aid future rational drug design targeting the receptor that could allow a more diverse range of therapeutic avenues. The findings may also clarify IP3R roles in cellular senescence and tumor suppression linked to selective vulnerability of cancer cells. Surprisingly, the study also clarifies a role of IP3Rs in the function of pathogenic unicellular organisms like Trypanosoma cruzi, the parasite of Chagas disease, and brucei, that causes African trypanosomiasis or sleeping sickness. The team identified an amino acid sequence in the leaflet that is conserved in parasites, suggesting structural insights that may assist in drug discovery for these devastating conditions.
Reference
- Kozo Hamada, Hideyuki Miyatake, Akiko Terauchi, and Katsuhiko Mikoshiba, "IP3-mediated gating mechanism of the IP3 receptor revealed by mutagenesis and X-ray crystallography", PNAS (Proceedings of the National Academy of Sciences of the United States of America)
Contact
Laboratory Head
Katsuhiko Mikoshiba
Laboratory for Developmental Neurobiology
RIKEN Brain Science Institute
Adam Phillips
RIKEN International Affairs Division
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Email: pr@riken.jp
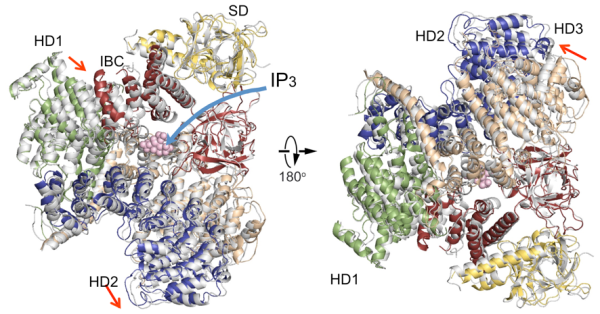
Comparison of IP3R cytosolic domain structures
This is a comparison of IP3R cytosolic domain structures in the absence of IP3 (colored) with one in the presence of IP3 (gray). Domain organization. Suppressor domain (SD), yellow; IP3-binding core (IBC), red; helical domain 1 (HD1), green; helical domain 2 (HD2), blue; helical domain 3, wheat.
