Oct. 9, 2019 Press Release Biology
How chromosomes change their shape during cell differentiation
The human genome is made up of 46 chromosomes, each of which has a length of about 100–200 million base pairs, base pairs being the building blocks of the DNA double helix. Even during interphase, the period in between the cell division phases, chromosomes are still tightly packed inside the cell nucleus. If one zooms in on each chromosome, a regular structural unit called the nucleosome is evident, which corresponds to a 146-base pair long strand of DNA wrapped around eight histone protein molecules. Until recently, no other regular structures beyond the nucleosomes were known.
Thanks to the emerging genomics-based technology called Hi-C (high-throughput chromosome conformation capture), however, we now know that there are regular structural units at the megabase scale, referring to millions of base pairs. It is now generally accepted that mammalian chromosomes are comprised of megabase-sized globular units called topologically associating domains (TADs), which are separated by boundaries, presumably in a beads-on-a-string manner. Further, multiple TADs assemble to form what are called A and B subnuclear compartments. TADs containing many active genes form A compartments, while TADs with few or no active genes form B compartments.
It is generally believed that TADs are stable units of the chromosomes and that their boundary positions do not change between cell types. By contrast, the organization of A/B compartments differs between cell types, meaning that the boundaries between them change during differentiation. However, nobody has ever observed changes in A/B compartments as they occurred.
Scientists from the RIKEN Center for Biosystems Dynamics Research have now observed A/B compartment changes in detail during the differentiation of mouse embryonic stem cells (mESCs). They discovered many genomic regions that switched compartments, either from A to B or vice versa, which, interestingly, correlated well with the genomic regions that switched their replication timing (the temporal order of genomic DNA replication) from early to late or vice versa, respectively. A to B compartment changes were accompanied by movements from the nuclear interior to the periphery and by gene repression, while B to A compartment changes were accompanied by movements from the nuclear periphery to the interior and by gene activation. These results strongly suggest that A/B compartment changes represent physical movements of portions of chromosomes within the 3D nuclear space, accompanied by changes in gene expression and replication timing.
Regarding the temporal relationship between the physical movements of chromosomes and changes in gene expression and replication timing, the research team found that genomic regions that switched from B to A compartment clearly did so one to two days prior to gene activation, and that the changes in replication timing were from late to early. This raised an intriguing possibility that compartment changes might be a prerequisite for gene activation and replication timing changes.
The team went on to characterize the features of genomic regions that changed A/B compartments. Compartments were found to change primarily by the shifting of A/B compartment boundaries, while the emergence of new compartments—for example the emergence of an A compartment within a stretch of B compartment or vice versa—was rare. Because compartment boundaries corresponded to a subset of TAD boundaries, they looked at how many TADs changed compartments and discovered that the majority of the changes affected single TADs.
Importantly, this single-TAD-level switching of compartments was confirmed in single cells by a method, called single-cell Repli-seq, which was recently developed by the research team to analyze DNA replication regulation genome-wide in single cells (note that replication timing correlates very well with A/B compartments). The team also found that A/B compartment profiles changed gradually but uniformly within a differentiating cell population, with the cells transiently resembling the epiblast-derived stem cell (EpiSC) state, an advanced form of stem cells compared to ESCs.
Taken together, the team’s finding suggests that A/B compartments change primarily by the relocation of single TADs facing the A/B compartment interface to the opposite compartment. “It is possible,” says Ichiro Hiratani, the leader of the group, “that the accumulation of these compartment switching events may reflect or represent changes in differentiation states such as from ESCs to EpiSCs.”
In this way, this study, published in Nature Genetics, explains how chromosomes undergo structural changes during cell differentiation. According to Hiratani, “Our study was the first to clearly demonstrate that changes in chromosome conformation preceded changes in DNA-based transactions such as gene expression and DNA replication timing. Intriguingly, chromosome conformation changes were regulated at the level of single TADs. We are eager to explore the basis of such single-TAD-level regulation of chromosomes and entertain the possibility of predicting DNA transactions based on preceding changes in chromosome structures.”
Reference
- Hisashi Miura, Saori Takahashi, Rawin Poonperm, Akie Tanigawa, Shin-ichiro Takebayashi, Ichiro Hiratani, "Single-cell DNA replication profiling identifies spatiotemporal developmental dynamics of chromosome organization", Nature Genetics, 10.1038/s41588-019-0474-z
Contact
Team Leader
Ichiro Hiratani
Laboratory for Developmental Epigenetics
RIKEN Center for Biosystems Dynamics Research
Jens Wilkinson
RIKEN International Affairs Division
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Email: pr [at] riken.jp
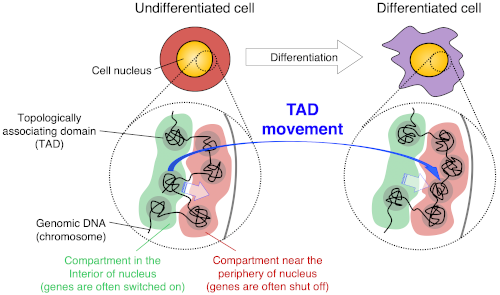
Schematic showing relationship between TADs and A/B compartments
