Nov. 10, 2017 Research Highlight Biology
The workings of a gene silencer
A protein that silences genes by packing DNA into an inactive form is guided into place by RNA
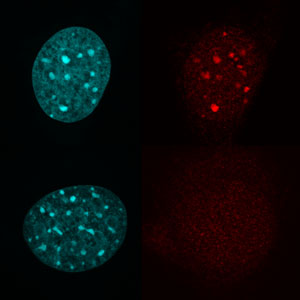 Figure 1: Stainings showing the location of DNA (left images) and trimethylation (right images) in cells with (top) and without (bottom) Suv39h1/2. They demonstrate that Suv39h1 or 2 is needed for trimethylation to occur. © 2017 RIKEN Cellular Memory Laboratory
Figure 1: Stainings showing the location of DNA (left images) and trimethylation (right images) in cells with (top) and without (bottom) Suv39h1/2. They demonstrate that Suv39h1 or 2 is needed for trimethylation to occur. © 2017 RIKEN Cellular Memory Laboratory
The molecular mechanisms guiding a key protein that regulates gene activity by modifying DNA have been clarified by RIKEN researchers1. They also discovered that RNA plays a crucial role in the process. These findings may enable researchers to artificially induce the process, allowing them to further dissect the mechanism and arrive at a full picture of its workings. Furthermore, they could also lead to a better understanding of how damage to an embryo’s genetic material can cause birth defects.
The proteins encoded by the two Suv39H genes are key players in modifying the protein–DNA complex to control gene activity. They chemically modify DNA-associated proteins called histones through a process known as methylation (Fig. 1). DNA domains tagged with these epigenetic marks are packaged into heterochromatin, a tightly bound form of DNA that silences genes by making them inaccessible.
The Suv39H proteins are known to bind to histones. Experiments on yeast in 2012 revealed they also bind to RNA to properly target histones for trimethylation. Many of the researchers involved in that study have now extended our understanding of these genes by showing that the same mechanism occurs in mammals.
Atsuko Shirai and Yoichi Shinkai of the RIKEN Cellular Memory Laboratory and co-workers used various protein analyses to demonstrate that Suv39h1 from mice binds to nucleic acids, particularly RNA, and that this binding helps to correctly target the protein. They also identified the RNA-binding domain of Suv39h1 and showed that it is distinct from the histone-binding domain; in other words, Suv39h1 binds to RNA and histones independently. Finally, by studying various Suv39h1 mutants, the researchers showed that binding to both RNA and histones is necessary for correct Suv39h1 targeting and heterochromatin assembly.
 Atsuko Shirai (left) and Yoichi Shinkai (right) have found that RNA plays a crucial role in guiding a protein that modifies DNA in order to silence genes. © 2017 RIKEN
Atsuko Shirai (left) and Yoichi Shinkai (right) have found that RNA plays a crucial role in guiding a protein that modifies DNA in order to silence genes. © 2017 RIKEN
“Suv39h1 is one of the most well characterized heterochromatin-regulating molecules,” says Shinkai. “If we can understand how Suv39h1 is targeted to accumulate at specific target loci, we may find that this mechanism generalizes to other epigenetic regulators.”
“Our study has clearly shown that RNA binding helps guide Suv39h1 to the correct location,” notes Shirai. “But it remains unclear whether RNA plays a role in the methylation reactions—we are now working on clarifying this.”
The team is also developing a system that uses Suv39h1 to induce heterochromatin formation at new regions in the genome. Shinkai hopes to artificially target Suv39h1 by introducing RNA guide molecules at novel locations, establishing a tool that will enable his team to dissect the mechanisms involved in trimethylation and heterochromatin formation and to distinguish the involvement of Suv39h1 in establishing and maintaining heterochromatin.
Related contents
References
- 1. Shirai, A., Kawaguchi, T., Shimojo, H., Muramatsu, D., Ishida-Yonetani, M., Nishimura, Y., Kimura, H., Nakayama, J. & Shinkai, Y. Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly. eLife 6, e25317 (2017). doi: 10.7554/elife.25317
