Dec. 20, 2019 Perspectives Biology
Why map every cell in the human body?
RIKEN is spearheading Asia’s efforts for the Human Cell Atlas, a push to understand the regulatory networks that make human cells unique.
 Image of various types of tissues. Credits; Skin: PASIEKA / Getty Images; Breast: Image Source / Getty Images; Lung: BeholdingEye / Getty Images
Image of various types of tissues. Credits; Skin: PASIEKA / Getty Images; Breast: Image Source / Getty Images; Lung: BeholdingEye / Getty Images
Piero Carninci: A highly standardized reference dataset of human cells is currently being created under a project called the Human Cell Atlas (HCA). This international consortium aims to create a common database consisting of millions of single-cell data points in order to map out all the cell types and their relationships. This will help better understand health and identify triggers that cause disease. The database aims to be open to the public to accelerate research for better medical research, diagnostics and drug discovery.1
I am part of the original HCA Organization Committee with Jay W. Shin, a team leader for the Laboratory for Advanced Genomics Circuit at the RIKEN Center for Integrative Medical Sciences. This committee consisted of 31 scientists from 10 countries and established the guidelines and scientific policies to make the HCA database an accessible resource to the global community. As of September 2019, more than 1,000 research institutes and 1,500 scientists from 60 countries and regions are now participating as members of the HCA.
As one of its key aims, the consortium is collecting data across broad ethnic and geographic regions to provide an equitable and globally representative picture of the human body. RIKEN is one of four Executive Offices (EOs) that will regularly coordinate general HCA activities, alongside the Wellcome Sanger Institute for the United Kingdom, the Broad Institute of MIT and Harvard for the United States, and the Karolinska Institutet for the European Union. RIKEN will act as the EO in Asia.
FANTOM synergy
RIKEN was perfectly positioned to take part in this project. It already has decades of experience mapping the gene expression and regulatory networks that define the different types of cells through its leadership of the international consortium working on FANTOM (Functional Annotation of the Mammalian Genome).
Among the many findings of the FANTOM projects, we cataloged the genomic regions critical in controlling the expression of RNAs, known as promoters and enhancers, and measured their activities across the major primary cells and tissues.2 The FANTOM database allowed us to understand the interplay of the regulatory proteins that defines a cell type, known as transcription factors. We also created an atlas of non-coding RNAs (ncRNAs) and revealed their importance in human genetics and gene control. In fact, in the current sixth edition of FANTOM (FANTOM6), we are aiming to understand the functions of long ncRNA.3
Because of this history, there is a natural synergy with the HCA and we will work closely with them to build the atlas of promoters and enhancers at a single-cell resolution, as well as to profile millions of single cells across many tissues across populations.
Toward the analysis of single cells
The HCA will start by sampling a comprehensive set of organs and tissues, followed by dissociating these samples into single cells or profiling gene expression within a tissue’s spatial context. The combined information will lead us to define the identity of each cell, based on the level of RNA, chromatin states and to a certain extent protein expression.
We will employ expert-guided curation and data-driven definitions to reach a consensus about the boundaries of how each cell should be classified by type and state, and this will create a common language for the global community.
RIKEN has started with a project to analyze blood samples from multiple ethnic groups in Asia including those in South Korea, Singapore (India, Malay, Chinese), and Japan funded by a grant from the Chan Zuckerberg Initiative (CZI). The project aims to compare cell-type compositions and RNA expression signatures in immune cells and cross-reference them to their sources’ genetic background, as well as to environmental factors, to identify the causal regulators of the immune system. Several bioinformaticians are on board and will work together with clinicians and ethicists to make the data meaningful and accessible to the community.
In addition to the CZI grant, RIKEN is spearheading a Single Cell Medical Network where samples from clinical collaborators in Japan will be profiled using the latest single-cell RNA sequencing and spatial technology.
In FANTOM5, we profiled large numbers of samples using CAP Analysis of Gene Expression (CAGE), which was developed at RIKEN. The CAGE method precisely determines where the gene is generated in our DNA, giving us a good sense of how the gene is controlled. However, in FANTOM5, we profiled bulk samples. Now, because of advancing technologies, we’re able to profile RNAs at a single-cell level using a modified version of the CAGE method that we recently developed.
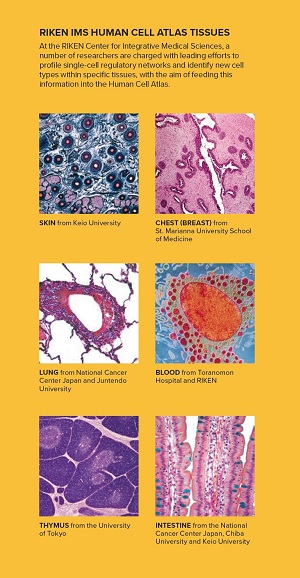 RIKEN IMS Human Cell Atlas Tissues: At the RIKEN Center for Integrative Medical Sciences, a number of researchers are charged with leading efforts to profile single-cell regulatory networks and identify new cell
types within specific tissues, with the aim of feeding this information into the Human Cell Atlas. Credits; Skin: PASIEKA / Getty Images; Breast: Image Source / Getty Images; Lung: BeholdingEye / Getty Images
RIKEN IMS Human Cell Atlas Tissues: At the RIKEN Center for Integrative Medical Sciences, a number of researchers are charged with leading efforts to profile single-cell regulatory networks and identify new cell
types within specific tissues, with the aim of feeding this information into the Human Cell Atlas. Credits; Skin: PASIEKA / Getty Images; Breast: Image Source / Getty Images; Lung: BeholdingEye / Getty Images
Jay W. Shin: Early in 2019, I led a team, together with Dr. Piero Carninci, to combine a microfluidic technique with CAGE to better and more systematically profile RNA transcripts at the single-cell level.
We used a microfluidic platform called C1, which isolates, processes and prepares individual cells for analysis, and we integrated a highly sensitive version of CAGE with this system, which we refer to as C1 CAGE. The team compared the C1 CAGE method with a standard protocol for profiling RNA's 5'ends in single cells and found it to be more sensitive, detecting more genes including non-coding RNAs.4
With the support of the HCA, we continue to develop and benchmark RNA profiling methods in single cells as one of our key contributions to the HCA community. With these new technologies we can measure individual RNA molecules within single cells, enabling us identify rare cell types or states in a sample with heterogeneous cell populations, which is particularly important for the interpretation of disease pathology in clinical samples of complex tissues.5
The ultimate aim of the HCA is to help treat diseases and improve diagnostics. There are many ways we can approach this. For example, it has become apparent that the risk of disease onset is usually linked to changes in the genome through single nucleotide polymorphisms, which can influence the functions and expression levels of the proteins that cause diseases. Analysis of RNA expression in single cells should shed light on exactly how each polymorphism affects expression levels and functions, helping us to vastly better understand diseases. By looking at deviations from the baselines set by the HCA, we should also be able to ascertain drug efficacy and safety with high precision.
The HCA will also help us look at diseases by revealing more cell subtypes. The brain, for example, has many subtypes of neurons, and some may only be discovered by looking at gene expression. In 2015, researchers at the Karolinska Institutet in Sweden discovered a much greater diversity of glial cells—a type of cell that supports and shields neurons—by looking at gene expression alone. We also know that certain subtypes of neurons are particularly affected in Parkinson’s disease, so to understand this type of disease, we must understand the brain’s cell types and the expression levels that make them unique.
In addition, cell states are important—for example, cells located near cancer cells may respond to signals from them and change states, becoming directly involved in cancer progression and metastasis. However, previous methods that relied upon the averaged values for groups of cells could not help us understand changes in cell states. The new single-cell CAGE could help us spot dynamic levels of RNA expression in cells in a disease state, as a means of early detection. For example, in Parkinson’s disease, cells that produce the neurotransmitter dopamine are lost in specific regions of the brain. As many as 70% of dopamine-producing cells are lost by disease onset, implying that these cells must show some markers earlier than that. We have already found that the level of a certain ncRNA in the blood increases in the early phase of Parkinson’s disease, which may lead to a means for early detection. Many more RNA and protein biomarkers are likely to be found in this way through the HCA.
Last but not least, a better understanding of cells will very likely help us when we differentiate induced pluripotent stem cells into specific cells to replace and repair damaged or defective tissues in the body.
Advances based on the HCA findings will likely be quick. The first cell atlas that will be published in a few years’ time will most likely be a draft of each healthy tissue and organ. New research can begin from the moment such an atlas is published, ranging from research into the similarities and differences between Asian and American lung cells, to differences by sex and age. It is likely that this research will also compare healthy tissues and organs characterized by the HCA with the cells of disease tissues and organs, opening up a whole range of exciting new insights.
References
- 1.Regev, A., et al. Science Forum: The Human Cell Atlas eLife 2017;6:e2704. doi: 10.7554/eLife.27041.001
- 2. Forrest, AR., et al. The FANTOM5. A promoter level mammalian expression atlas. Nature 507, 462–470 (2014). doi: 10.1038/sdata.2017.112
- 3. Hon, CC., et al. An atlas of human long non-coding RNAs with accurate 5' ends. Nature 543, 199–204 (2017). doi: 10.1038/nature21374
- 4. Kouno, T., et al. C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution. Nature 10, 360 (2019). doi: 10.1038/s41467-018-08126-5
- 5. Mereu, E., et al. Benchmarking Single-Cell RNA Sequencing Protocols for Cell Atlas Projects. doi: 10.1101/630087
About the Researcher
Piero Carninci, Deputy Director of the RIKEN Center for Integrative Medical Sciences (IMS)

After arriving at RIKEN in 1995, Carninci played a critical role in many key projects, including the FANTOM Consortium. In 2018, he became deputy director of the IMS. He also leads the Laboratory for Single Cell Technologies, which is describing samples as populations of single cells classified by RNAs. He is an organizing committee member of the HCA.
Jay W. Shin, Team Leader at the RIKEN Center for Integrative Medical Sciences (IMS)

Shin studied computer science at Boston College in the United States and gained a PhD in life sciences at Swiss Federal Institute of Technology in Zürich, Switzerland. Today, he leads RIKEN’s Laboratory for Advanced Genomics Circuit, co-leads the FANTOM6 consortium and is an organizing committee member of the HCA.
