Apr. 4, 2024 Research Highlight Biology
Analysis of gene expression helps bring order to complex skin condition
By studying RNA taken from the blood and skin, researchers have brought greater order to a complex skin disease
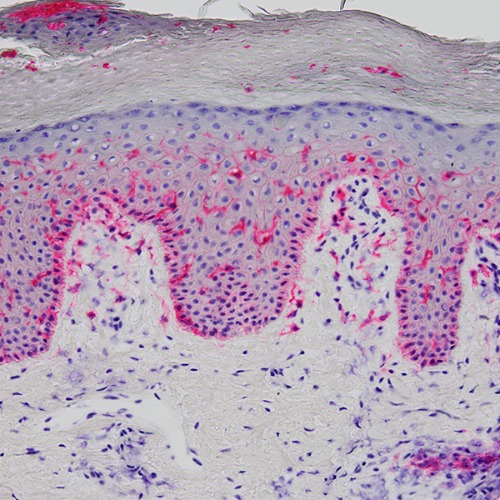
Figure 1: A histological image of the skin from a patient with atopic dermatitis. The skin biospecimen was immunohistochemically stained for CD1a, a marker of Langerhans cells, which are antigen-presenting cells involved in immune surveillance. RIKEN researchers have uncovered associations between phenotypes and endotypes of atopic dermatitis based on an analysis of RNA in blood and skin. © 2024 RIKEN Center for Integrative Medical Sciences
RIKEN biologists have found links between different manifestations of a common skin disease and RNA in skin and blood samples taken from patients1. In the future, this could help to prescribe treatments that are tailored to individuals.
One of the most common forms of eczema, atopic dermatitis causes the skin to become itchy, dry or edematous.
It is a complex condition that depends on both genetics and environmental factors. For example, a person whose parents both have atopic dermatitis has a higher risk of developing the disease compared to when only one parent or neither parent has it. But other factors besides genetics also play a role.
“The frequency of atopic dermatitis increases as societies become more industrialized, because the disease is linked with the environment,” says Aiko Sekita of the RIKEN Center for Integrative Medical Sciences (IMS).
Atopic dermatitis can also display a range of symptoms. For example, it can give rise to redness in skin (known as erythema) or it can cause raised bumps on the skin (papulation).
There are also indications that it is more than a disease of the skin, but involves other organs as well.
But there hasn’t been an effective way of classifying atopic dermatitis to bring order to this confusion. Being able to better classify atopic dermatitis would help understand how and what causes it and also help prescribe suitable treatments in specific cases.
Now, Sekita, Haruhiko Koseki, also of the IMS, and co-workers have compared different clinical presentations of atopic dermatitis with RNA taken from skin and blood samples from 115 patients and 14 healthy controls.
Based on their results, they were able to develop a model that can predict features associated with different forms of the skin condition from gene-expression data. In particular, they were able to differentially characterize the erythema and papulation forms of atopic dermatitis based on different immunological signatures.
In addition, the team analyzed data from blood samples taken from 30 atopic-dermatitis patients over a year. They found that patterns of disease activity, such as remission and exacerbation, were associated with patterns of gene-expression changes.
The findings provide a useful framework for future investigations of the complex disease. “Our results have made it clear that RNA data is useful for interpreting this complex disease,” notes Sekita.
The study also has implications for studying other complex diseases. “A key message is that complex diseases should be considered at a system level in biology,” says Sekita. “Researchers may need to analyze both the specific organs where a disease manifests itself but also other organs that can communicate with each other.”

Haruhiko Koseki (left), Aiko Sekita (right) and co-workers have brought greater order to atopic dermatitis, a complex skin disease, based on an analysis of RNA in blood and skin. © 2024 RIKEN
Related content
Rate this article
Reference
- 1. Sekita, A., Kawasaki, H., Fukushima-Nomura, A., Yashiro, K., Tanese, K., Toshima, S., Ashizaki, K., Miyai, T., Yazaki, J., Kobayashi, A. et al. Multifaceted analysis of cross-tissue transcriptomes reveals phenotype–endotype associations in atopic dermatitis. Nature Communications 14, 6133 (2023). doi: 10.1038/s41467-023-41857-8
