Sep. 29, 2006 Research Highlight Biology
Expanding the genetic code
Researchers add a third pair of ‘letters’ to DNA’s natural alphabet
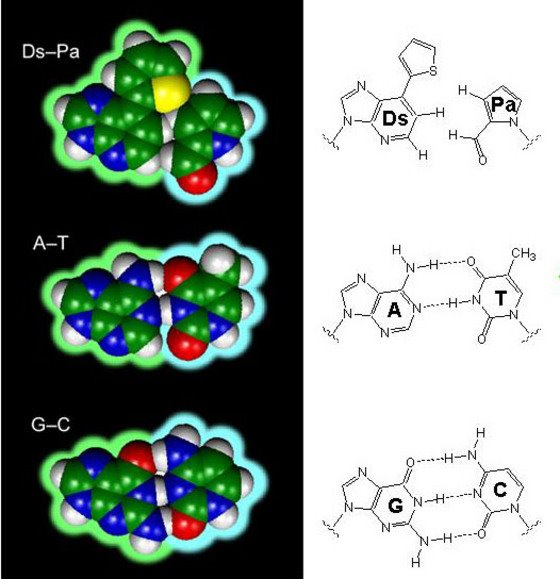 Figure 1: The unnatural base pair system, Ds-Pa, compared with the natural base pairs, A-T and G-C.
Figure 1: The unnatural base pair system, Ds-Pa, compared with the natural base pairs, A-T and G-C.
Molecular biologists at the RIKEN Genomic Sciences Center in Yokohama have constructed a synthetic pair of nucleotide bases. These bases can be incorporated into the genetic compounds of DNA and RNA, and replicated and transcribed by the standard enzymes found in cells.
The new pair, an imidazo-pyridine and a pyrrole-carbaldehyde known as Ds and Pa respectively, add two more alternatives to the existing four ‘letters’ of the genetic code—guanine (G) and cytosine (C), and adenine (A) and thymine (T). The research team believes these ‘unnatural’ bases can be used to introduce molecular groups with novel and interesting properties into specific positions in DNA and RNA. Already the group has attached fluorescent tags to RNA by means of the new bases.
The unnatural base pair may even open the way towards building novel amino acids into ‘unnatural’ proteins, according to project leader, Ichiro Hirao.
In a report of their work in Nature Methods 1, the researchers describe how Ds and Pa are driven by the affinity of the surrounding water molecules for each other to pack tightly together in a complementary fashion in what is known as a hydrophobic interaction. In contrast, the natural bases of A-T and G-C pair via an electrostatic interaction called hydrogen bonding (Fig. 1).
Although more than 50 such unnatural base pairings have been reported in the literature, this is the first that can be routinely manipulated by standard enzymes. The researchers showed in the laboratory that DNA incorporating Ds and Pa can be replicated and amplified with a high degree of fidelity using the polymerase chain reaction—although the process involves using modified energy-rich phosphate compounds. They also demonstrated that the unnatural base pair can be transcribed from DNA into RNA, which opens the possibility of using it to construct novel proteins or RNA-based compounds.
The researchers are now exploring the development of functional DNA and RNA with different active molecular groups attached via the new base pairs. “We are also trying to establish a translation system using unnatural base pairs for site-specific incorporation of various unnatural amino acids into proteins,” says Hirao.
In addition, Hirao wants to try to incorporate the Ds-Pa base pair into a living organism, such as a bacterium. The organisms would be inherently safe, Hirao insists, because they cannot produce all of the compounds they require to function. For example, the researchers would have to supply some of the compounds required in replication and translation.
References
- 1. Hirao, I., Kimoto, M., Mitsui, T., Fujiwara, T., Kawai, R., Sato, A., Harada, Y. & Yokoyama, S. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nature Methods 3, 729–735 (2006). doi: 10.1038/nmeth915
