Feb. 9, 2007 Research Highlight Biology
How plants work with sulfur
Researchers unravel a process essential to human wellbeing

Molecular biologists from the RIKEN Plant Science Center in Yokohama have identified a key genetic transcription factor which increases the efficiency of uptake and use of sulfur by plants in sulfur-deficient environments.
The work is significant as all plants need sulfur for growth. Many agricultural species—particularly the Brassicas, such as canola, broccoli and cabbages—require sulfur fertilization for maximum crop yields.
Sulfur also is an essential nutrient for animals, including humans, as it is an active part of two amino acids of critical importance to the structure of proteins. While plants can take up sulfur from the soil in the form of sulfates, animals can only acquire sulfur by eating plants or other animals.
The research could lead to more efficient use of fertilizers, according to team leader, Hideki Takahashi. This would reduce over-application and consequent pollution of waterways. Plants also use sulfur to make defensive compounds. One major group of these produced in Brassicas, the glucosinolates, is well known for protecting humans against cancer. “Our work may help us engineer increased glucosinolate production in Brassica crops,” Takahashi says.
Boosting sulfur uptake
In response to sulfur deficiency, plants are able to boost their ability to take up sulfates from the soil, to release and transport sulfur within themselves, and to break down their sulfur-rich defense chemicals, such as glucosinolates, and recycle the sulfur they contain. This response is highly coordinated across the whole plant. So the research team reasoned that it might be initiated by a single critical regulator, perhaps a transcription factor, the essential link in the process of transcribing genes into messenger RNAs that generate the proteins and enzymes necessary to maintain the cellular function. Such a regulatory factor could simultaneously switch on the production of several different proteins necessary to the adjustment of the plant’s metabolism in response to the sulfur supply shortage.
Finding and verifying the critical factor
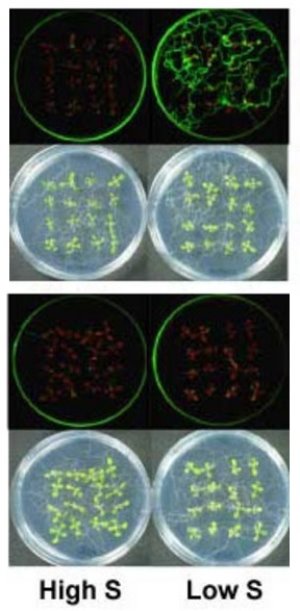 Figure 1: Arabidopsis plants carrying the normal SLIM1 gene fluoresce under low sulfur conditions (upper). Plants with a mutant version of SLIM1 do not fluoresce (lower).
Figure 1: Arabidopsis plants carrying the normal SLIM1 gene fluoresce under low sulfur conditions (upper). Plants with a mutant version of SLIM1 do not fluoresce (lower).
In a recent paper in The Plant Cell 1, Akiko Maruyama-Nakashita and researchers from Takahashi’s team describe how they were able to track down this critical factor—which they called SLIM1—in the thale cress, Arabidopsis thaliana. They did so by carefully fashioning a gene which produces a fluorescent protein under the control of the most important sulfate transporter—the ion transporting protein responsible for sulfate uptake—generated in response to low sulfur conditions (Fig. 1). This genetically modified plant fluoresces when this sulfate transporter is present and active under low sulfur conditions.
Using this ‘sulfur indicator’ plant, the team searched for those individuals which did not fluoresce under low sulfur conditions, suggesting they possessed a mutant form of SLIM1, unable to stimulate the production of the sulfate transporter. The mutants they discovered led them back to the SLIM1 gene itself. They then set out to demonstrate SLIM1’s involvement in the other parts of the coordinated response.
First, they were able to verify SLIM1’s overall involvement in the process. They did so by adding and activating the normal SLIM1 gene to plants which carried the mutant form of the gene. This restored the typical response to sulfur-deficient environments. Further, the team demonstrated how widespread and important the mechanism is in plants, by using the rice plant version of the SLIM1 gene to compensate for its inactivity in Arabidopsis mutants.
Then, the group used gene chip technology to determine which genes SLIM1 switches on or activates, and hence which compounds and biochemical pathways it affects. Gene chips are able to show which genes are active in cell extracts, and which are not. The researchers compared gene activity under sulfur-deficient conditions in the roots of plants with normal and with mutant SLIM1 genes.
The results indicate that under sulfur-deficient conditions SLIM1 is responsible for switching on genes for sulfate transporters and enzymes that break down glucosinolates, and for switching off genes for the enzymes that produce the glucosinolates in the first place. This work could be cross-referenced and verified by studying the compounds that are produced and accumulated in the mutant plants when they are grown in sulfur-deficient conditions.
The researchers were also able to link SLIM1 back to a gene already named in the Arabidopsis genome. They showed that the mutants of the SLIM1 gene fall into the region of the Arabidopsis genome which encodes one of a family of six transcription factors known collectively as ethylene-insensitive-like (EIL) genes. The SLIM1 gene is, in fact, EIL3. They verified that this is the only member of the family specialized for the sulfur response by using each of the six EIL genes in turn to try to compensate for SLIM1 in the mutant plants. Only EIL3 was able to do so.
Clearly, not all the impacts of SLIM1 are caused directly by its protein product. The transcription factor sets off a cascade of downstream biochemical reactions. So the research team is now looking more closely at the SLIM1-mediated response, says Takahashi, to track down the additional protein factors necessary for the metabolic response in plants.
One goal is to engineer plants adapted to low-sulfur environments. Another, says Maruyama-Nakashita, is to explore the potential for producing a wider range of edible crop plants that incorporate cancer-protective glucosinolate compounds.
References
- 1. Maruyama-Nakashita, A., Nakamura, Y., Tohge, T., Saito, K. & Takahashi, H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. The Plant Cell 18, 3235–3251 (2006). doi: 10.1105/tpc.106.046458
About the Researcher
Hideki Takahashi, Akiko Maruyama-Nakashita

Hideki Takahashi was born in 1967, and graduated from Kyoto University in 1990 with a major in biochemical engineering. From 1990 to 1993, he worked as a research associate at Idemitsu Petrochemical, and then earned his doctorate in pharmaceutical sciences from Chiba University in 1998. From 1998 to 2000, Takahashi served as a postdoctoral fellow at Chiba University and the Carnegie Institution of Washington, US. In 2000, he was appointed as the leader of the Plant Nutrition and Basal Metabolism Research Team at the RIKEN Plant Science Center.
Akiko Maruyama-Nakashita was born in 1971, and graduated from Tohoku University in 1995 with a major in biology. In 1997, she earned her masters degree in biology, and obtained her doctorate in biology in 2000, both from Tohoku University. After working as a postdoctoral fellow at Chiba University for a year, she joined Takahashi’s laboratory at RIKEN in 2001 as a research scientist.
