Mar. 16, 2007 Research Highlight Biology
Inheriting a defective genome
Researchers begin to solve a genetic puzzle
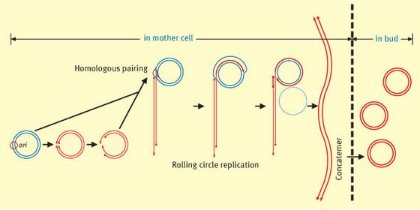 Figure 1: The proposed model of rolling-circle replication that produces the linear head-to-tail array of multiple mitochondrial genomic units known as concatemers. Mol. Cell. Biol. /American Society for Microbiology /27/1143 (2007).
Figure 1: The proposed model of rolling-circle replication that produces the linear head-to-tail array of multiple mitochondrial genomic units known as concatemers. Mol. Cell. Biol. /American Society for Microbiology /27/1143 (2007).
Using yeast, molecular biologists at RIKEN have put forward a new model of how a normal genome is inherited in mitochondria, and thus how a faulty genome can be preferentially passed on to future generations. The research provides insight into the mechanics of mitochondrial genetics in yeast, and may have relevance to mammals.
Mitochondria provide energy for all cell functions, and contain their own double-stranded DNA which encodes several of the enzymes and other components critical to their role. This mitochondrial DNA (mtDNA) is separate from the nucleus, and comes packaged in a circular chromosome typical of micro-organisms, such as bacteria. Thus, when the mitochondria divide asexually in cells, the method of replication differs from that of nuclear DNA.
Generally, all the thousands of copies of the mtDNA within cells are the same. Sometimes, however, defective copies of mtDNA with large deletions arise and can accumulate in specific tissues, with disastrous health results in humans. The team from RIKEN’s Discovery Research Institute in Wako studied how this happens in yeast.
In earlier work, the researchers demonstrated that replication of mtDNA in yeast involves the formation of concatemers, multiple copies of the genomic DNA linked head to tail in a line. The concatemers were shown to be produced by a process known as rolling-circle replication (Fig. 1). When passed on to the next generation, the concatemers are broken into individual copies each of which forms into a circular chromosome.
In the conventional scheme of replication, an RNA primer is synthesized and initiates the unidirectional process at a particular sequence called a replication origin. In the latest work reported in Molecular and Cellular Biology1 the team found that both DNA strands are cut at the replication origin known as ori5. One of the cut strands serves as a primer onto which connected copies are synthesized using intact circular mtDNA as a template (Fig. 1).
The researchers suggest that the mutant genomes contain greater numbers than usual of the replication origin sequence. The resulting mutant concatemers contain more copies of the smaller defective genome than is the case with the normal mtDNA chromosome. Hence, greater numbers of the mutant genome are passed on to the next generation, and almost all progeny contain mitochondria with the mutant genome.
“We hope our work will cause people to rethink the basic mechanism of mitochondrial replication,” say team members, Feng Ling and Takehiko Shibata. “We now want to extend our work into mammalian systems.”
References
- 1. Ling, F., Hori, A. & Shibata, T. DNA recombination-initiation plays a role in the extremely biased inheritance of yeast [rho-] mitochondrial DNA that contains the replication origin ori5. Molecular and Cellular Biology 27, 1133–1145 (2007). doi: 10.1128/MCB.00770-06
