Mar. 21, 2008 Research Highlight Biology
The key to controlling DNA copying
Researchers use beamlines at RIKEN’s SPring-8 Center to glean new insights into how replication is regulated
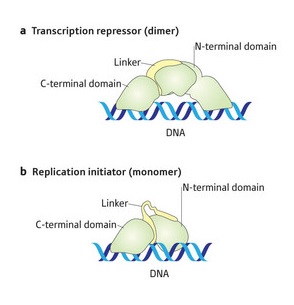 Figure 1: Schematics of the structural difference in the dimeric (upper) and monomeric (lower) RepE protein.
Figure 1: Schematics of the structural difference in the dimeric (upper) and monomeric (lower) RepE protein.
Molecular biologists in Japan have unraveled structural details of a key molecular process which regulates DNA replication—the basis of reproduction.
The researchers used as their model the F-plasmid associated with sexual reproduction in the bacterium Escherichia coli—a small piece of genetic material which operates independently to the chromosome. They believe, however, that while the fine detail of control of DNA replication may differ in other locations and organisms, the general pattern will be the same.
Previous work has shown that the protein RepE plays an essential role in regulating the copying of the F-plasmid, ensuring there are only one to two plasmids in each individual bacterial cell. Single RepE molecules, or monomers, join together to generate a double form, or dimer, which is the predominate form in the cell. The dimer binds to the promoter/operator of the repE gene preventing its transcription and repressing replication, whereas the monomer binds to a small repeated DNA sequence to initiate replication.
The conversion between the two forms is known to be mediated by the DnaK system of three chaperone proteins, but the structural details of how this might occur were unclear because no dimeric structure had been published for RepE. In a recent paper published in the Proceedings of the National Academy of Sciences 1, the researchers from Kyoto University and RIKEN detail how they used the beamlines at RIKEN’s SPring-8 Center in Harima to generate the first structure of the RepE dimer and then compared it with the monomer.
They successfully prepared the dimer under high salt conditions and obtained crystals of the complex with the repE operator DNA. The monomer consists of two independent regions at each end known as domains—a C-terminal domain and an N-terminal domain. They are joined by a linker region. In the domains the conformation, or 3-D disposition of atoms to each other, remains constant in both monomer and dimer. But the orientation of the domains to each other is strikingly different between the two forms, exhibiting structural changes in the linker regions connecting them (Fig. 1).
The researchers found that sequences in the linker regions provide convenient binding points for the molecular chaperones that regulate the splitting of the dimer into two monomers thereby activating DNA replication. “Now we will try to prepare the RepE-chaperone complex,” says team-member, Akira Nakamura from Kyoto University. “It should provide good information on the detail of the activation process.”
References
- 1. Nakamura, A., Wada, C. & Miki, K. Structural basis for regulation of bifunctional roles in replication initiator protein. Proceedings of the National Academy of Sciences USA 104, 18484–18489 (2007). doi: 10.1073/pnas.0705623104

