Mar. 15, 2013 Research Highlight Biology
The complexity of regulated development in plants
The herculean task of defining the physical interactions and locations of hundreds of Arabidopsis proteins involved in targeted protein degradation is underway
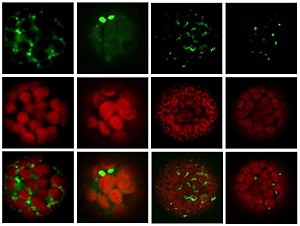 Figure 1: Four Arabidopsis FBX proteins (green, top and bottom rows) preferentially accumulate in speckled structures in the cytoplasm of protoplasts in leaves, outside of chloroplasts (red, middle and bottom rows). © 2012 Kuroda et al.
Figure 1: Four Arabidopsis FBX proteins (green, top and bottom rows) preferentially accumulate in speckled structures in the cytoplasm of protoplasts in leaves, outside of chloroplasts (red, middle and bottom rows). © 2012 Kuroda et al.
In most living organisms, growth and development are controlled by selective modification of the lifespans of particular proteins. This mechanism is especially prevalent in plants, allowing rapid moderation of gene expression. Even the relatively streamlined Arabidopsis genome encodes more than 1,400 components of ubiquitin ligase complexes—molecular machines that are each able to single out a specific type of protein for degradation while sparing tens of thousands of others. However, the selectivity with which these hundreds of components assemble to form complexes has yet to be defined.
A team led by Minami Matsui from the Plant Functional Genomics Research Group at the RIKEN Plant Science Center has now begun to explore the diversity of the largest group of plant ubiquitin ligases—the SCF complexes—in Arabidopsis. Each plant SCF complex comprises four canonical components, two of which are an ASK protein and an FBX protein. The Arabidopsis genome encodes an estimated 897 FBX and 21 ASK proteins. In their most recent work, the researchers investigated the locations of selected FBX and ASK proteins and their ability to interact with each other1.
Knowing the locations of proteins is equally as important as understanding their interaction, says Yuki Yanagawa from the research team. “After all, even if two proteins are capable of physical interaction, that’s irrelevant in physiological terms if the proteins are found in different tissues or subcellular compartments.”
The team used a yeast-based assay to map the physical affinities of each of the 341 Arabidopsis FBX proteins for each of the 19 ASK proteins. More than half the FBX proteins didn’t interact with any of the ASK proteins tested, suggesting other components, SCF complexes or post-translational modifications might be needed to facilitate certain FBX–ASK interactions. A complementary experiment suggested, however, that the original assay may also have underestimated the capacities of certain FBX proteins to interact with ASKs.
The authors then analyzed a database of gene expression data collected from 79 Arabidopsis tissue types to identify groups of FBX and ASK proteins with similar tissue-specific expression profiles. This revealed that SCF complexes are particularly likely to be active in reproductive tissues. They subsequently examined the subcellular locations of 17 FBX proteins by fluorescent tagging (Fig. 1).
“This is a promising start to understanding how many distinct SCF complexes can be formed from different combinations of FBX and ASK proteins,” says Matsui. “Eventually, we’ll be able to pinpoint the roles of hundreds of SCF complexes in plant growth and development.”
References
- 1. Kuroda, H., Yanagawa, Y., Takahashi, N., Horii, Y. & Matsui M. A comprehensive analysis of interaction and localization of Arabidopsis SKP1-LIKE (ASK) and F-Box (FBX) Proteins. PLoS One 7, e50009 (2012). doi: 10.1371/journal.pone.0050009
