Apr. 12, 2013 Research Highlight Biology
Stopping the rot
Genomic analysis reveals the genetic basis of pathogenicity in crop-blighting fungi
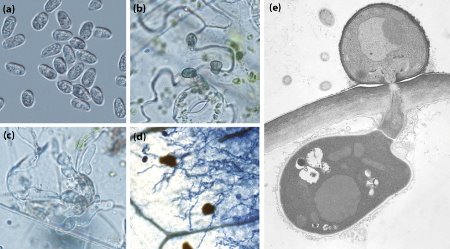 Figure 1: Life-cycle stages of Colletotrichum orbiculare. (a) Germination, (b) penetration of plant cells, (c) fungal proliferation, (d) development of fungal filaments associated with plant death. (e) Fungal infection of plant cell, showing fungal penetration into the cell and the formation of an infection vesicle. Reproduced from Ref. 1 © 2012 P. Gan et al.
Figure 1: Life-cycle stages of Colletotrichum orbiculare. (a) Germination, (b) penetration of plant cells, (c) fungal proliferation, (d) development of fungal filaments associated with plant death. (e) Fungal infection of plant cell, showing fungal penetration into the cell and the formation of an infection vesicle. Reproduced from Ref. 1 © 2012 P. Gan et al.
The fungi Colletotrichum orbiculare and C. gloeosporioides represent major threats to crops around the world, responsible for blight and post-harvest rot in plants as diverse as melons, strawberries, mangoes, coffee and leaf crops. Both of these fungi are ‘hemibiotrophs’—first growing parasitically on living plant tissue then killing the host and feeding on the remains. Ken Shirasu, Pamela Gan and colleagues of the Plant Immunity Research Group at the RIKEN Center for Sustainable Resource Science have now sequenced the entire genomes of these two fungi and predicted their gene functions to identify the genetic basis for their hemibiotrophic pathogenicity1.
Fungal pathogenicity in plants is known to occur through biochemical interaction between the fungus and host involving genetically encoded secreted proteins. The researchers found through their genomic analysis of C. orbiculare and C. gloeosporioides that both species, while having quite different genomes, exhibit similar ‘expanded’ gene classes that are not augmented in non-hemibiotrophic fungi. These included genes for digestive enzymes that degrade plant cell walls, for chitin-binding proteins that may help protect the fungal cell wall from plant defenses, and for the synthesis of secondary metabolites likely to be related to pathogenicity. Genes for small secreted proteins (SSPs), which are thought to facilitate fungal colonization, were also similarly expanded.
Shirasu’s team also studied gene expression in C. orbiculare during infection of the model plant Nicotiana benthamiana (Fig. 1). They found distinct patterns in the timing of expression over the life-cycle of the fungus: SSPs, secondary metabolite synthesis enzymes and some chitin-binding proteins were found to be active early in host colonization, whereas digestive enzymes were most concentrated during plant death. The timing of secondary metabolite synthesis indicates that fungal secondary metabolites are not predominantly toxins as previously assumed, but instead may contribute to the manipulation of host growth during colonization. The presence of high levels of SSPs at an early stage suggests these are also important for colonizing the host.
The findings highlight the importance of different gene classes for all stages of hemibiotrophic fungus development, and the set of genes identified provides a crucial base for future investigation into pathogen activity. Ultimately, this research could lead to control strategies for these and other fungi, such as the development of crops with genes that provide resistance to secreted fungal proteins. “Knowing which plant resistance genes are effective against which pathogens aids in informing the deployment of rapid and targeted crop management strategies,” says Gan.
References
- 1. Gan, P., Ikeda, K., Irieda, H., Narusaka, M., O’Connell, R.J., Narusaka, Y., Takano, Y., Kubo, Y. & Shirasu, K. Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytologist 197, 1236–1249 (2013). doi: 10.1111/nph.12085
