Jul. 4, 2014 Research Highlight Chemistry Physics / Astronomy
X-ray imaging reveals a complex core
A novel structural analysis scheme using multiple x-ray techniques probes the internal three-dimensional structure of macromolecular complexes
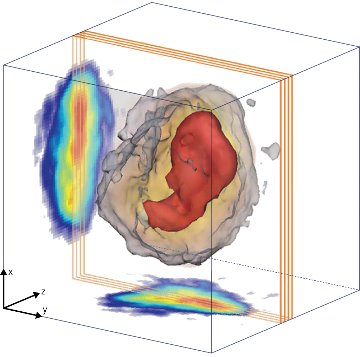 Figure 1: The complete three-dimensional structure of an RNA interference (RNAi) microsponge. Reproduced, with permission, from Ref. 1 © 2014 M. Gallagher-Jones et al.
Figure 1: The complete three-dimensional structure of an RNA interference (RNAi) microsponge. Reproduced, with permission, from Ref. 1 © 2014 M. Gallagher-Jones et al.
Macromolecular complexes composed of self-assembling proteins and nucleic acids hold promise for a wide range of applications, including drug delivery, sensing and molecular electronics. Scientists have developed a broad array of x-ray scattering techniques for characterizing the shape and surface morphology of these complexes, but probing their internal structure remains a challenge.
Changyong Song and colleagues from the RIKEN SPring-8 Center and RIKEN Advanced Institute for Computational Science have now devised a structural analysis scheme based on complementary advanced x-ray techniques that has allowed them to peer inside an RNA macromolecular complex for the first time1.
Although knowledge of their shape and morphology is useful, it is the internal structure of macromolecular complexes that holds the secret to their formation and disassembly. RNA interference (RNAi) microsponges, for example, are self-assembled from small interfering RNA molecules to form macromolecular complexes that are capable of silencing gene expression.
The researchers set out to explore the internal structure of RNAi microsponges using a combination of x-ray techniques at the SPring-8 synchrotron radiation facility. They first performed nondestructive diffraction imaging experiments using a coherent x-ray beam. The experiments yielded high-resolution diffraction patterns that the researchers used to reconstruct projection images of RNAi microsponges (Fig. 1). The results suggest that all RNAi microsponges have a high-density core that occupies approximately 15% of the projected area.
The team then performed single-shot imaging experiments using femtosecond x-ray pulses produced by the SACLA x-ray free electron laser (XFEL) facility at SPring-8. The use of intense, extremely short x-ray pulses helped compensate for the weak scattering from the RNAi microsponges. The studies revealed that irrespective of their size, all of the microsponges shared almost the same electron density distribution, suggesting that the high-density core evolves gradually through a dynamic compaction of RNA strands.
Finally, the researchers conducted small-angle scattering experiments using the XFEL. The results further confirmed that the core–shell structure is a generic feature of RNAi microsponges.
The x-ray structural analysis scheme is nondestructive, providing deeper penetration into target materials and enhanced resolution limited only by the wavelength of x-rays used. The scheme is also highly versatile because, unlike many other analysis schemes, no prior information about the sample is required to complete the structural analysis.
“X-ray diffraction is a versatile technique for understanding atomic structures, and with XFELs the technique has become even more dexterous,&uo; says Song. “Multimodal imaging combining both the SPring-8 and SACLA facilities now provides a route to the atomic world.”
References
- 1. Gallagher-Jones, M., Bessho, Y., Kim, S., Park, J., Kim, S., Nam, D., Kim, C., Kim, Y., Noh, D. Y., Miyashita, O. et al. Macromolecular structures probed by combining single-shot free-electron laser diffraction with synchrotron coherent X-ray imaging. Nature Communications 5, 3798 (2014). doi: 10.1038/ncomms4798
