May 15, 2015 Research Highlight Physics / Astronomy
Electrons reveal the hidden structure of proteins
An innovative crystallography technique reveals the charge distribution in protein structures
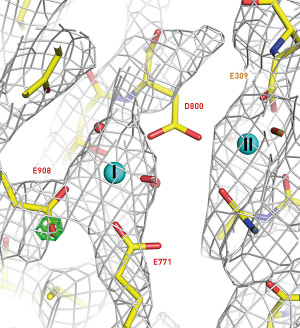 Figure 1: A Coulomb potential map showing the distribution of charge (cages) around the Ca2+ binding sites of Ca2+-ATPase. Reproduced, with permission, from Ref. 1 © 2015 K. Yonekura et al.
Figure 1: A Coulomb potential map showing the distribution of charge (cages) around the Ca2+ binding sites of Ca2+-ATPase. Reproduced, with permission, from Ref. 1 © 2015 K. Yonekura et al.
An electron crystallography technique developed by RIKEN and the University of Tokyo makes it possible to probe hidden details of small three-dimensional protein crystals as a potent new tool for studying the function and structures of biological macromolecules1.
The atomic structure of crystalline materials is routinely determined by a technique called x-ray crystallography, in which the scatter of x-rays is used to identify the location of atoms in the crystal. This method can be used to reveal the structures of many proteins, but for macromolecular complexes and proteins that occur in cell membranes, it is often very difficult to obtain crystals large enough and yielding clear enough diffraction patterns to be able to determine their structure using even the most intense x-rays.
Electron crystallography is based on a similar principle but uses a beam of electrons instead of x-rays and has the advantage of being 100,000 times more strongly scattered by organic molecules. This makes it possible to use much smaller and thinner crystals, which are all that are available for many biologically important proteins. Electron crystallography can also resolve charge states, revealing critical information about protein function. However, the electron beam can damage the sample, and collecting and analyzing data from crystals a few molecular layers thick is challenging.
Koji Yonekura from the RIKEN SPring-8 Center and Chikashi Toyoshima from the University of Tokyo led a collaborative study to develop an electron crystallography method that overcomes these limitations through the use of a new diffractometer and novel data collection and processing techniques. “As no technology was available for electron crystallography of such thin crystals,” says Yonekura, “we had to develop both the hardware and software, in many cases from scratch.”
To reduce radiation damage, the new diffractometer operates at cryogenic temperatures. To obtain useful structural information from the weak electron scattering signals, the diffraction spots produced by scattered electrons are recorded and stacked while the crystal sample is rotated. A new data analysis package then produces a precise atomic model that includes charge information.
The team tested the technique on ultrathin crystals of two common enzymes, Ca2+-ATPase and catalase, producing ‘Coulomb potential’ maps showing the charge states of ion and amino acid residues at the enzymes’ active sites (Fig. 1).
These maps, note Yonekura, show key aspects of the working mechanism of the biological macromolecules. “Using this technique we have resolved the charge states of ions and amino acids—this information cannot be obtained using other techniques.”
References
- 1. Yonekura, K., Kato, K., Ogasawara, M., Tomita, M. & Toyoshima, C. Electron crystallography of ultrathin 3D protein crystals: Atomic model with charges. Proceedings of the National Academy of Sciences USA 112, 3368–3373 (2015). doi: 10.1073/pnas.1500724112
