May 22, 2015 Research Highlight Biology
Mapping a metabolic master switch
The crystal structures of two signaling proteins yield promising leads for better treatments for metabolic diseases
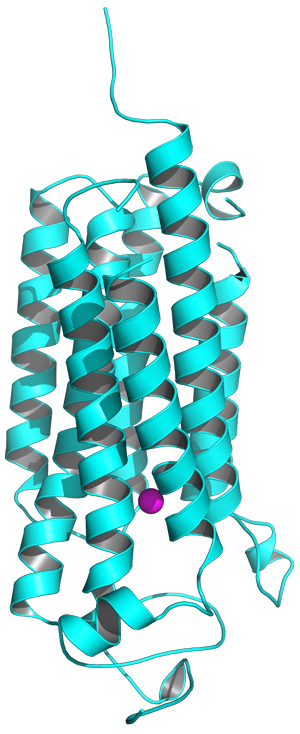 Figure 1: The structure of the AdipoR2 receptor, showing the intracellular (bottom) and extracellular (top) components, and the position of the zinc ion (purple). Reprinted by permission from Macmillan Publishers Ltd: Nature (Ref.1), copyright 2015
Figure 1: The structure of the AdipoR2 receptor, showing the intracellular (bottom) and extracellular (top) components, and the position of the zinc ion (purple). Reprinted by permission from Macmillan Publishers Ltd: Nature (Ref.1), copyright 2015
A detailed structural analysis of a pair of unusual proteins involved in metabolism by a RIKEN-led research team gives valuable insight into a newly discovered class of receptors1. The results could help guide the development of drugs that more effectively target the physiological processes underlying diabetes and obesity.
The protein adiponectin is secreted into the bloodstream by fat cells and subsequently binds to receptors in the muscle, liver and other tissues to initiate physiological processes such as sugar and fat metabolism. Adiponectin triggers these responses through two different receptors, AdipoR1 and AdipoR2, which have recently emerged as appealing drug targets.
“AdipoR1 and AdipoR2 are key molecules associated with metabolic syndrome,” explains Shigeyuki Yokoyama, director of the RIKEN Structural Biology Laboratory. Indeed, scientists have already uncovered at least one drug candidate that can act on these receptors and has been shown to improve health and lifespan in diabetic and obese rodents.
AdipoR1 and AdipoR2 both penetrate the cell membrane. The external portion binds to adiponectin, and the internal portion interacts with cellular machinery (Fig. 1). These receptors contain the same seven-transmembrane-helical architecture as an important class of proteins called G-protein-coupled receptors (GPCRs), which transmit signals critical in a variety of disease states. However, the adiponectin receptors are essentially ‘backward’ compared to GPCRs—the domain that faces into the cell in one family faces outside the cell in the other. This suggests that these proteins may differ considerably in their mechanism.
To better understand these receptors, Yokoyama’s group teamed up with the researchers from the University of Tokyo who originally cloned AdipoR1 and AdipoR2, Takashi Kadowaki and Toshimasa Yamauchi. The joint team was able to generate extremely detailed structural models for both receptors, and learn more about how they interact with adiponectin.
Differences large and small
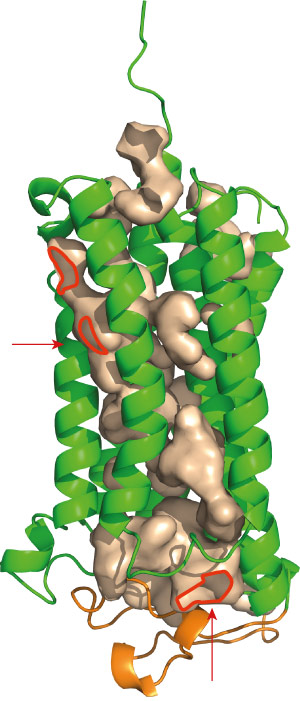 Figure 2: Both AdipoR1 (above) and AdipoR2 maintain immobilized zinc ions within cavities (red) that lead deep into the protein. Reprinted by permission from Macmillan Publishers Ltd: Nature (Ref.1), copyright 2015
Figure 2: Both AdipoR1 (above) and AdipoR2 maintain immobilized zinc ions within cavities (red) that lead deep into the protein. Reprinted by permission from Macmillan Publishers Ltd: Nature (Ref.1), copyright 2015
It is very difficult to convert highly dynamic membrane-bound proteins into the orderly crystals required for x-ray analysis, but Yokoyama and his colleagues were able to successfully employ some tricks that had previously proved useful for analyzing the GPCR structure. They trimmed off short segments of AdipoR1 and AdipoR2 that are particularly poorly structured, and used an antibody fragment that binds tightly to the exterior of the receptor to lock the protein into a more stable state. The resulting crystals were of sufficient quality to yield x-ray structures with a resolution of higher than 0.3 nanometers.
The data showed that the two receptors are much more than just ‘backward GPCRs’. “We expected that the structures of AdipoR1 and AdipoR2 would be very different from those of GPCRs,” says Yokoyama. “However, the structures we determined are completely new and surprising.” Indeed, a search of other known protein structures revealed no obvious relatives to the two proteins, although the two receptors were highly similar to each other.
Among the most distinctive features of the adiponectin receptors is a cavity containing a zinc ion, situated deep within the inner membrane of the cell (Fig. 2). Yokoyama and his colleagues subsequently generated a series of mutants lacking the amino acids that hold this zinc ion in place. Although these mutations appeared to destabilize the AdipoR1 protein, they had a more notable effect on AdipoR2: altering just one of the four amino acids reduced adiponectin-mediated activation of gene expression, and multiple mutations essentially eliminated this activity. The researchers hypothesize that AdipoR2 may employ the zinc ion to directly catalyze the production of intermediary biomolecules that stimulate gene activation, and note that the large cavity surrounding this ion may facilitate the entry and exit of substrates.
Each of these receptors initiates a distinct series of downstream signaling events. Although the two proteins are largely similar in structure, the research team noted striking differences in the amino acid content of a particular intracellular segment. “This may be relevant to the difference in the downstream signaling pathways between these receptors,” notes Yokoyama. The outward-facing adiponectin-binding sites, on the other hand, were highly similar, and the researchers identified multiple loops in the protein structure that may contribute to the recognition of this molecule.From structure to function
Researchers have been able to develop a drug that can selectively bind and activate both AdipoR1 and AdipoR2, even in the absence of a detailed structural map. With the new structural details obtained by Yokoyama’s team, however, it should be possible to generate even more effective therapeutic agents that are designed based on a more in-depth understanding of the atomic-scale interactions between adiponectin and its receptors. Yokoyama and his colleagues are now investigating how these structures can be translated into accurate functional models of adiponectin signaling. “One of our goals is to understand the activation mechanisms of AdipoR1 and AdipoR2,” says Yokoyama. “Therefore, we would like to solve the structures of AdipoR1 and AdipoR2 in a complex with adiponectin.”
These findings will be of significant interest to the research community as well as to scientists pursuing clinical applications. The mammalian adiponectin receptors have counterparts even in distantly related forms of life, including plants and yeast, suggesting that this newly characterized class of proteins has played a critical role throughout the course of evolutionary history. Although plants do not grapple with fat and blood sugar regulation, they appear to exploit a related signaling process for self-defense. “Plants secrete an antifungal protein, osmotin, which causes cell death in the yeast species Saccharomyces cerevisiae by acting on its version of AdipoR,” says Yokoyama. Intriguingly, osmotin can also exert anti-obesity effects by acting on human AdipoR1 and AdipoR2, and his group will also be characterizing how these receptors interact with this protein in comparison to adiponectin.
References
- 1. Tanabe, H., Fujii, Y., Okada-Iwabu, M., Iwabu, M., Nakamura, Y., Hosaka, T., Motoyama, K., Ikeda, M., Wakiyama, M., Terada, T. et al. Crystal structures of the human adiponectin receptors. Nature 520, 312–316 (2015). doi: 10.1038/nature14301
About the Researcher
Shigeyuki Yokoyama

Shigeyuki Yokoyama was born in Tokyo, Japan, in 1953. He received his Bachelor of Science and PhD degrees from the University of Tokyo in 1975 and 1981, respectively, followed by five years of postdoctoral work. Yokoyama became an associate professor in 1986 and a professor in 1991 at the University of Tokyo’s Department of Biophysics and Biochemistry. In 1993, he joined RIKEN as chief scientist of the RIKEN Cellular Signaling Laboratory, later becoming project director of the Protein Research Group at the RIKEN Genomic Sciences Center. Between 2008 and 2013, Yokoyama directed the RIKEN Systems and Structural Biology Center and since April 2013 has been heading the RIKEN Structural Biology Laboratory as distinguished senior scientist.
