Jun. 10, 2016 Perspectives Medicine / Disease
A healthy balance
RIKEN IMS aims to improve medical therapy by getting a clearer understanding of the delicate equilibrium maintained by the body in times of health.
 The RIKEN Center for Integrative Medical Sciences is advancing our understanding of the factors that contribute to disease to help people maintain homeostasis—a condition of internal stability—and good health. © mattpail/ RooM/ Getty
The RIKEN Center for Integrative Medical Sciences is advancing our understanding of the factors that contribute to disease to help people maintain homeostasis—a condition of internal stability—and good health. © mattpail/ RooM/ Getty
The Ancient Greek physician Hippocrates was one of the first to associate homeostasis—a condition of internal stability—with health. A state of well-being, he explained, consists of a balance between the four ‘humors’ produced by the body: blood, phlegm, yellow bile and black bile. Any excess, deficiency or change in the quality of these fluids results in disease.
While our description of homeostasis has changed over the past millennia, the concept remains central to our understanding of life and death. The body constantly makes adjustments to maintain a stable internal environment, such as dropping blood sugar levels after a candy binge, or generating heat in response to cold temperatures. These internal stabilizing systems keep the body in check, preventing disease and ultimately death.
Both environmental and genetic factors can disrupt the body’s ability to maintain stability and fight disease. The common skin disorder atopic dermatitis, for example, has long been known to run in families. However, while many genes may contribute to a child’s risk of developing the condition, exposure to toxins, irritants and pollutants are equally important in triggering a reaction1.
The RIKEN Center for Integrative Medical Sciences (IMS) is advancing our understanding of the internal and external factors that contribute to disease to help people maintain homeostasis and good health. Established in 2013, through a merger between the former RIKEN Research Center for Allergy and Immunology and the RIKEN Center for Genomic Medicine, the IMS combines a long history of research into the body’s critical artillery for protecting itself against foreign threats, the immune system, with the development of large-scale genome studies to uncover the sources of instability that lie within.
DNA banks
Significant technological advances over the past decade have led to deeper insights into the agents underpinning or upsetting homeostasis. The IMS has been developing and utilizing state-of-the-art technologies for the analysis of information within the human genome.
Following the success of the Human Genome Project in sequencing the entire human genome in 2003, it became possible to study genomic variations within and between entire populations. Scientists hypothesized that differences in the code could explain observable differences in physical traits, and even offer clinically relevant information about disease susceptibility. The development of faster, cheaper and more accurate genome sequencing technologies made these studies possible.
In 2002, researchers at RIKEN published one of the first genome-wide association studies (GWAS) looking for single-nucleotide differences between the genomes of a group of people2. This study compared the genomes of about 100 Japanese heart-attack patients and around 650 healthy individuals. Since then, close to 2,500 GWAS studies have associated more than 16,000 variations in DNA lettering with diseases ranging from macular degeneration, to type 2 diabetes and multiple sclerosis.
In 2003, the former RIKEN Center for Genomic Medicine joined a national BioBank project to collect hundreds of thousands of blood samples from Japanese citizens for further genome studies. Analyses of these samples have since identified more than 34 billion single-nucleotide variations and 170 genes associated with specific diseases, 20 of which respond to drugs.
Kazuhiko Yamamoto at the IMS has been using the BioBank’s genome data for the past decade to search for the genetic basis of rheumatoid arthritis, a complex autoimmune disease characterized by swelling and pain in the joints. In 2014, he collaborated with a large international research team in a GWAS of 100,000 individuals of European and Asian descent3. The team connected 101 nucleotide links in the DNA chain with a risk of developing rheumatoid arthritis—every single-letter switch representing a lead for a potential treatment. An extensive drug search found that approved drugs for rheumatoid arthritis already target proteins expressed by 27 genes linked with an increased risk of the disease, and approved cancer drugs overlap with 2 genes. The approach taken by Yamamoto is a powerful, unbiased and data-driven route to drug discovery.
Annotated DNA
 Figure 1: Makoto Arita’s laboratory at the RIKEN Center for Integrative Medical Sciences uses advanced mass spectrometry to characterize the metabolites present in a cell, tissue or organism. © 2016 Makoto Arita, RIKEN Center for Integrative Medical Sciences
Figure 1: Makoto Arita’s laboratory at the RIKEN Center for Integrative Medical Sciences uses advanced mass spectrometry to characterize the metabolites present in a cell, tissue or organism. © 2016 Makoto Arita, RIKEN Center for Integrative Medical Sciences
While sequencing the genome is an important first step to understanding many diseases, scientists still have a lot to learn. Every cell in the body contains an identical copy of DNA, but a neuron’s interpretation of the genetic code varies considerably from that of a cardiac muscle cell, for example. One of the keys to this diversity lies in how DNA is packaged.
Advanced sequencing technologies can now observe the hustle and bustle around the genome coordinating genetic expression at specific moments in time. In 2012, IMS researcher Katsuyuki Shiroguchi improved on the accuracy of an RNA sequencing technique by developing a digital barcode method for counting the RNA in a sample, and has been refining the technique at the IMS for single-cell analysis4. Systematic tools have also been developed for characterizing and quantifying the proteins (proteomics) and small-molecule chemicals (metabolomics) present in a sample. Makoto Arita’s laboratory, for example, uses advanced mass spectrometry to analyze the structure and function of many types of lipids synthesized and degraded by the body (Fig. 1). These lipid metabolites play an important role in regulating inflammation and maintaining tissue homeostasis5.
Supercomputers and advanced computational systems have also allowed vast amounts of information to be stored and processed within manageable timeframes. And live cell and in vivo imaging tools can capture the vibrant detail of life in motion, even in full color.
Functional analysis
To observe how genetic and environmental factors control homeostasis at a systemic level, researchers rely on another well-known experimental tool—the model organism. Animal models have been used to study the effects of knocking out crucial genes, to analyze disease pathogenesis, and to test the therapeutic or toxic effects of various compounds. To investigate the role of environmental factors, researchers at the IMS frequently use germ-free organisms, which are born and maintained in sterile, isolated chambers.
But animal models can be poor representations of human biology and disease. To address this problem, Fumihiko Ishikawa at the IMS has developed a method for studying the behavior of human cells, tissues and organisms in ‘humanized’ mice. The technique involves transplanting human cells into mouse models with weak immune systems to ensure that the foreign cells are not rejected, instead growing and differentiating in their host.
Using this method, Ishikawa has differentiated human stem cells isolated from umbilical cord blood and introduced to germ-free mice into mature blood cells, including cell types essential to the immune system such as dendritic cells and lymphocytes6. Humanized mice facilitate the study of human immunity in vivo, from examining our response to pathogens and tumors to testing vaccines against infectious diseases and cancers.
Environmental factors
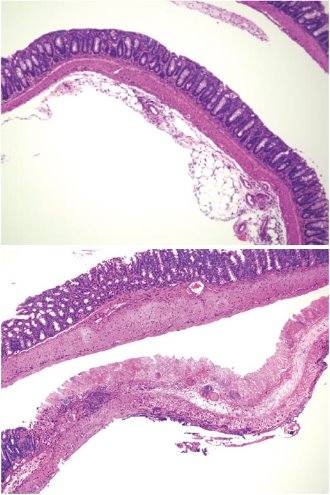 Figure 2: Mouse models of colon inflammation treated with a mixture of 17 bacterial strains had healthier guts (upper) than those who did not receive the treatment (lower). © Reprinted by permission from Macmillan Publishers Ltd: Nature Ref. 9, copyright 2013
Figure 2: Mouse models of colon inflammation treated with a mixture of 17 bacterial strains had healthier guts (upper) than those who did not receive the treatment (lower). © Reprinted by permission from Macmillan Publishers Ltd: Nature Ref. 9, copyright 2013
From genomics and its many -omics derivatives to computational science and animal experiments, researchers at the IMS have become adept at consolidating the latest tools and techniques to trace the origins of stability in the human body.
This integrated approach is particularly relevant in the field of immunology. Shohei Hori, for example has clarified the mechanism by which regulatory T cells prevent other lymphocytes from attacking themselves7, a suicidal behavior that if unsuppressed can cause a range of autoimmune diseases such as type 1 diabetes, thyroiditis and inflammatory bowel disease.
IMS researchers, including Hiroshi Ohno, Sidonia Fagarasan and Kenya Honda have further identified how the self-protective function of regulatory T cells is shaped by microbes living in the gut. Their research has revealed the critical role of interactions between the body and its gut microbes in maintaining immune homeostasis; any breakdown in communication could cause inflammation8.
Honda took this research a step further by colonizing the guts of germ-free mice with bacterial strains taken from either a healthy human or patients of ulcerative colitis. Seventeen of the introduced bacterial strains from the healthy individuals were found to induce anti-inflammatory molecules and T regulatory cell activity9, while a cocktail of bacterial strains from patients of ulcerative colitis induced pro-inflammatory immune cells10 in the mouse guts. The bacterial strains from the healthy humans were subsequently fed to mouse models of colon inflammation. The diet-treated diseased mice showed milder symptoms, such as less severe diarrhea and reduced colon shortening, and lower mortality rates (Fig. 2).
Clinical harvest
Modern medicine and research is one of the main reasons people today live longer than any generation before them. But the challenge for society will be to ensure that longevity progresses alongside physical, mental and social well-being. This will require more research into homeostasis and its role in human health and disease, an endeavor that will require collaboration between researchers and clinicians.
In 2016, RIKEN launched a project called the Innovation Hub, led by Kazuhiro Sakurada together with Hiroaki Kitano, Tatsuhiko Tsunoda and Haruhiko Koseki at the IMS, to bring these two groups closer together. In partnership with four hospitals across Japan, it will collect and analyze blood samples from patients with rheumatoid arthritis and atopic dermatitis. Using machine-learning and artificial intelligence to process the incoming data, the project will hopefully lead to new markers for diagnosing and eventually even predicting the onset of disease. The IMS is expected to play a pivotal role in this project.
A fragmented approach to medical science is no longer sustainable. The IMS’s model of integrating research follows in the footsteps of the great father of modern medicine, Hippocrates.
References
- 1. Yasuda, T., Fukada, T., Nishida, K., Nakayama, M., Matsuda, M., Miura, I., Dainichi, T., Fukuda, S., Kabashima, K., Nakaoka, S. et al. Hyperactivation of JAK1 tyrosine kinase induces stepwise, progressive pruritic dermatitis. The Journal of Clinical Investigation 126, 2064-2076 (2016). doi: 10.1172/JCI82887
- 2. Ozaki, K., Ohnishi, Y., Iida, A., Sekine, A., Yamada, R., Tsunoda, T., Sato, H., Sato, H., Hori, M., Nakamura, Y. & Tanaka, T. Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nature Genetics 32, 650–654 (2002). doi: 10.1038/ng1047
- 3. Okada, Y., Wu, D., Trynka, G., Raj, T., Terao, C., Ikari, K., Kochi, Y., Ohmura, K., Suzuki, A., Yoshida, S. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014). doi: 10.1038/nature12873
- 4. Shiroguchi, K., Jia, T. Z., Sims, P. A. & Xie, X. S. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proceedings of the National Academy of Sciences USA 109, 1347–1352 (2012). doi: 10.1073/pnas.1118018109
- 5. Endo, J., Sano, M., Isobe, Y., Fukuda, K., Kang, J. X., Arai, H. & Arita, M. 18-HEPE, an n-3 fatty acid metabolite released by macrophages, prevents pressure overload-induced maladaptive cardiac remodeling. The Journal of Experimental Medicine 211, 1673–1687 (2014). doi: 10.1084/jem.2013201
- 6.Shultz, L. D., Saito, Y., Najima, Y., Tanaka, S., Ochi, T., Tomizawa, M., Doi, T., Sone, A., Suzuki, N., Fujiwara, H. et al. Generation of functional human T-cell subsets with HLA-restricted immune responses in HLA class I expressing NOD/SCID/IL2rγnull humanized mice. Proceedings of the National Academy of Sciences USA 107, 13022–13027 (2010). doi: 10.1073/pnas.1000475107
- 7.Hori, S. Lineage stability and phenotypic plasticity of Foxp3⁺ regulatory T cells. Immunological Reviews 259, 159–172 (2014). doi: 10.1111/imr.12175
- 8.Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., Nakanishi, Y., Uetake, C., Kato, K., Kato, T. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). doi: 10.1038/nature12721
- 9.Atarashi, K., Tanoue, T., Oshima, K., Suda, W., Nagano, Y., Nishikawa, H., Fukuda, S., Saito, T., Narushima, S., Hase, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013). doi: 10.1038/nature12331
- 10.Atarashi, K., Tanoue, T., Ando, M., Kamada, N., Nagano, Y., Narushima, S., Suda, W., Imaoka, A., Setoyama, H., Nagamori, T. et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015). doi: 10.1016/j.cell.2015.08.058
About the Researcher
Tadashi Yamamoto

Tadashi Yamamoto is director of the RIKEN Center for Integrative Medical Sciences. He is interested in the intracellular signaling pathways and gene expression activated in response to extracellular signals. His current research focuses more specifically on the coordinated mechanisms controlling mRNA stability.
