Jan. 20, 2017 Research Highlight Chemistry
Lighting up drug discovery
A simple molecular tag offers a new way to study potential drug targets
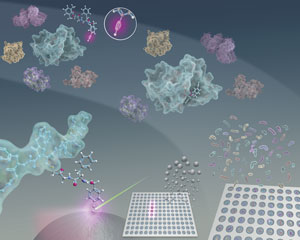 Figure 1: The tiny alkyne tag (inset at top of image) on a small molecule enables to find the modified fragment of target protein (light blue structures) in a complex mixture. The Raman signal from the alkyne, amplified by silver nanoparticles (sphere at bottom left of image), was used to optically screen the small-molecule-modified fragments. © 2017 Mikiko Sodeoka, RIKEN Chief Scientist Laboratory
Figure 1: The tiny alkyne tag (inset at top of image) on a small molecule enables to find the modified fragment of target protein (light blue structures) in a complex mixture. The Raman signal from the alkyne, amplified by silver nanoparticles (sphere at bottom left of image), was used to optically screen the small-molecule-modified fragments. © 2017 Mikiko Sodeoka, RIKEN Chief Scientist Laboratory
Some proteins can be most uncooperative. Some refuse to form nice crystals for analysis by x-ray crystallography, others are too insoluble for nuclear magnetic resonance (NMR), while still others—including some of great interest for their role in disease—can be isolated only in trace amounts.
Researchers at RIKEN have developed a new way to study these problematic proteins and their interactions with small molecules, a discovery that could greatly benefit drug discovery research1.
When studying how a protein interacts with a potential drug, it is essential to find out where the drug binds on the protein. For proteins that cannot be analyzed by x-ray or NMR, researchers try to covalently bond a drug-like molecule to the protein, break up the protein and then use mass spectrometry to pick out the relevant protein fragment based on its predicted mass. But it can be difficult to identify the protein fragment of interest from such a complex cocktail of protein pieces.
Now, Mikiko Sodeoka from the RIKEN Synthetic Organic Chemistry Laboratory and her colleagues, in collaboration with Katsumasa Fujita at Osaka University, have found a way to overcome this problem. They tagged small molecules and then analyzed them using a technique called Raman spectroscopy, which is similar to infrared spectroscopy in that it involves using light to excite molecules and examining how they vibrate.
Molecules known as alkynes contain a triple bond between two carbon atoms; this bond produces a telltale signal in the Raman spectrum. Thus, by incorporating an alkyne into a small molecule that binds to a protein, the protein fragment of interest from a complex mixture can be quickly identified using Raman spectroscopy (Fig. 1). Since alkynes are small, they rarely interfere with the interaction between the small molecule and the protein.
“All the equipment we used is commercially available,” Sodeoka says, “But we made considerable effort to optimize and automate the analysis conditions.” One of the team’s most important optimizations was the addition of silver nanoparticles, since they powerfully amplify the alkyne’s Raman signal.
The researchers demonstrated the potential of their technique by using it to identify the drug-binding site of a protein called cathepsin B, an anticancer drug target. They were able to pick out the protein from the highly complex protein mixture produced by rupturing a cell.
The method is not limited to the analysis of proteins, Sodeoka notes. “We expect it could contribute to various research fields such as proteomics, epigenetics and metabolomics.”
Related contents
References
- 1. Ando, J., Asanuma, M., Dodo, K., Yamakoshi, H., Kawata, S., Fujita, K. & Sodeoka, M. Alkyne-tag SERS screening and identification of small-molecule-binding sites in protein. Journal of the American Chemical Society 138, 13901–13910 (2016). doi: 10.1021/jacs.6b06003
