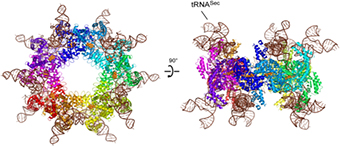
Researchers led by Dr. Shigeyuki Yokoyama from the Structural Biology Laboratory at the Center for Life Science Technologies have determined the crystal structure of the Sec synthase, SelA, responsible for the Ser-to-Sec conversion in bacteria. They determined the structures of SelA alone and in a complex with tRNASec.
They find that SelA is a homodecamer in which the 10 subunits form a pentamer of dimers. This pentamer forms a ring-shaped structure and binds 10 tRNASecmolecules. Each tRNASec interacts with four subunits from the two neighboring dimers, and this interaction critically depends on the pentagonal ring architecture.
The SelA N-terminal domain recognizes the tRNASec-specific D-arm structure, thereby discriminating Ser-tRNASec from Ser-tRNASer. Moreover, the large cleft created between two SelA dimers accommodates the Ser-tRNASec 3′-terminal region.
The authors conclude that the bacterial and archaeal/eukaryotic Sec synthases have completely different structures and divergent residues for substrate recognition and catalysis, which provides evidence for a convergent evolution of two Sec synthesis systems present in nature.
The study is published in the journal Science (doi: 10.1126/science.1229521)
