Dec. 17, 2014 Press Release Biology Medicine / Disease
Predicting antibiotic resistance
Treating bacterial infections with antibiotics is becoming increasingly difficult as bacteria develop resistance not only to the antibiotics being used against them, but also to ones they have never encountered before. By analyzing genetic and phenotypic changes in antibiotic-resistant strains of E. coli, researchers at the RIKEN Quantitative Biology Center (QBiC) in Japan have revealed a common set of features that appear to be responsible for the development of resistance to several types of antibiotics.
The study published in Nature Communications shows that resistance emerges through mutations that converge on similar physical changes in the bacteria. Quantifying these changes by measuring the expression of a small number of genes can be useful in predicting a bacteria’s response to a given antibiotic, and knowing which genes are important may contribute to the development of new ways to prevent resistance.
To perform this complex genetic and phenotypic analysis, Shingo Suzuki, Takaaki Horinouchi, and Chikara Furusawa first used a technique called laboratory evolution to create 44 strains of E. Coli, each resistant to one of 11 different antibiotics. Then they examined how each of these resistant strains responded to 25 antibiotics they had never encountered. The tests showed that most strains had developed resistance to several of the 25—a phenomenon called cross-resistance—even when these antibiotics worked differently from the one used to generate the resistance. The team also found that in the case of two classes of antibiotics, cross-susceptibility developed—bacteria that become resistant to one type became more vulnerable to the other.
The researchers reasoned that similar alterations in gene expression might be one cause of cross-resistance. To test this hypothesis, they identified the changes in gene expression for each of the resistant strains using microarray analysis. Then, they combined this information with the resistance, cross-resistance, and cross-susceptibility data from some strains to make a simple linear model that did a very good job predicting the resistance and susceptibility patterns of the remaining strains. Furusawa notes that, “these high-precision predictions were possible using a small number of genes, making this a powerful way to describe a bacteria’s phenotype and its expected response to antibiotics.”
The researchers also looked for fixed mutations that might link the development of resistance across antibiotics. For example, they found that almost all strains had fixed mutations affecting a particular multidrug efflux pump—a pump that bacteria use to expel unwanted molecules. However, one of the main findings was that although the bacteria showed similar changes in expression patterns, these often resulted from different changes in the genome, and frequently from a combination of several different mutations. Furusawa speculates that, “this type of convergent evolution may be a key factor that drives the development of antibiotic resistance.”
The cross-susceptible classes of antibiotics were found to have gene expressions and fixed mutations that did not overlap at all. Strains resistant to aminoglycoside antibiotics, for example, showed mutations and down-regulation of genes that while effective in blocking aminoglycoside antibiotics, also resulted in less effective multidrug efflux pumps. This explains why these strains became more susceptible to all the other antibiotics—the bacteria could not send them out of their cells.
Understanding the common factors that result in antibiotic resistance could help combat this growing problem. As Furusawa reflects, “by making it possible to quantitatively determine what genes contribute to the development of antibiotic resistance, this research could lead to new methods for blocking acquisition of resistance and to the development of new antibiotic compounds.”
Reference
- Shingo Suzuki, Takaaki Horinouchi, and Chikara Furusawa, "Prediction of antibiotic resistance by gene expression profiles", Nature Communications, 2014, doi: 10.1038/ncomms6792.
Contact
Team Leader
Chikara Furusawa
Laboratory for Multiscale Biosystem Dynamics
Computational Biology Research Core
RIKEN Quantitative Biology Center
Jens Wilkinson
RIKEN Global Relations and Research Coordination Office
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Email: pr@riken.jp
Colorful E.Coli
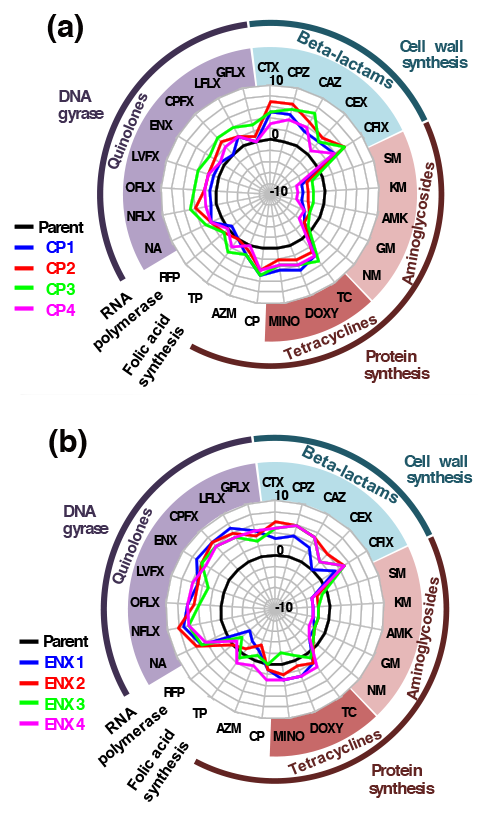
Two examples of cross-resistance and cross-susceptibility that developed after laboratory evolution.
