Apr. 6, 2017 Press Release Biology
New technique helps researchers determine how stem cells differentiate
Stem cell differentiation can now be seen thanks to a combination of machine learning and microfabrication techniques developed by scientists at the RIKEN Quantitative Biology Center in Japan. The results, published in PLOS One, followed the differentiation of human mesenchymal stem cells (MSCs) which are easily obtained from adult bone marrow.
MSCs have proven to be important for regenerative medicine and stem cell therapy because they can potentially repair many different types of organ damage, as they have the ability to differentiate into various cell types including bone, muscle and fat. Depending on the way the cells are grown the results can be quite different and so controlling differentiation is an important goal.
Observing MSC differentiation under different conditions is an essential step in understanding how to control the process. However, this has proved challenging on two fronts. First, the physical space in which the cells are grown has a dramatic impact on the results, causing significant variation in the types of cells into which they differentiate. Studying this effect requires consistent and long lasting spatial confinement. Second, classifying the cell types which have developed through manual observation is time consuming.
Previous studies have confined cell growth with fibronectin on a glass slide. The cells can only adhere and differentiate where the fibronectin is present and are thus chemically confined. However, this procedure requires high technical skill to maintain the confinement for an extended period of time. To overcome this, the first author of the study, Nobuyuki Tanaka, decided to look for a new way to confine them. Using a simple agarose gel physical confinement system, he found that he could maintain them for up to 15 days. Tanaka says, “It was wonderful to be able to do this, because agarose gel is a commonly used material in biology laboratories and can be easily formed into a micro-cast in a PDMS silicone mold.”
He continues, “The advantage of this system is that once the PDMS molds are obtained the user only needs agarose gel and a vacuum desiccator to create highly reproducible micro-casts.” The vacuum pump pulls the agarose gel into the mold. He explains, “We provided the protocol to our coauthors at ETH Zurich and they performed the agarose micro-casting and conducted the stem cell differentiation study. Stem cells were captured in the micro-structures and their differentiation was controlled under the captured condition.”
Tanaka’s paper also describes an automated cell type classification system, using machine learning, which reduces the time and labor needed to analyze cells. “Combined together, these tools give us a powerful way to understand how stem cells differentiate in given conditions.”
According to Yo Tanaka, leader of the Laboratory for Integrated Biodevice, where the research was conducted, “We hope this will break down the barriers that have hindered research in this area so far and help to establish harmony between biologists and engineers. The focus of engineers has traditionally been to develop new technologies, but scientists prefer to use well established technologies. However, if our newly developed technology is simple enough it can spread rapidly, this is our goal.”
Reference
- Nobuyuki Tanaka, Tadahiro Yamashita, Asako Sato, Viola Vogel, and Yo Tanaka, "Simple agarose micro-confinement array and machine-learning-based classification for analyzing the patterned differentiation of mesenchymal stem cells", PLOS ONE, doi: 10.1371/journal.pone.0173647
Contact
Unit Leader
Yo Tanaka
Research Scientist
Nobuyuki Tanaka
Laboratory for Integrated Biodevice
Laboratory for Synthetic Biology
Cell Design Research Core
RIKEN Quantitative Biology Center
Jens Wilkinson
RIKEN International Affairs Division
Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
Email: pr@riken.jp
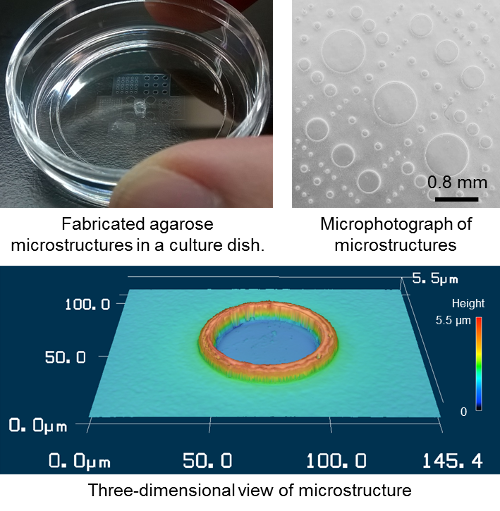
Images of the microstructures
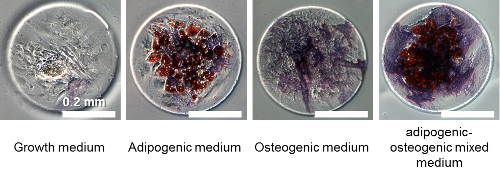
Differentiated stem cells in various media
