Aug. 18, 2017 Research Highlight Biology
Mapping disease genes to immune cells
Gene variants tied to rheumatoid arthritis and other diseases can now be traced to particular faulty immune cells
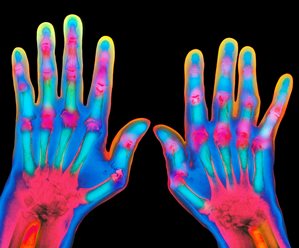 Figure 1: Researchers demonstrate a new way to identify gene variants associated with complex diseases and a dysregulated pathway in faulty immune cells by applying the approach to rheumatoid arthritis. © SCIENCE PHOTO LIBRARY
Figure 1: Researchers demonstrate a new way to identify gene variants associated with complex diseases and a dysregulated pathway in faulty immune cells by applying the approach to rheumatoid arthritis. © SCIENCE PHOTO LIBRARY
A valuable analytical tool developed by RIKEN researchers could help to reveal when multiple genes are all working through a common pathway in specific immune cells that underpin complex diseases such as diabetes or heart disease1.
To demonstrate the tool’s effectiveness, the scientists applied it to DNA datasets from rheumatoid arthritis sufferers and confirmed the importance of immune dysregulation through a cell-signaling protein called tumor necrosis factor (TNF). Anti-TNF drugs are some of the best-selling medicines in the world—a fact that Kazuyoshi Ishigaki of the RIKEN Center for Integrative Medical Sciences says confirms the “robustness of our pipeline.” As such, he adds, “It’s reasonable to expect that our pipeline can be applied to other diseases.”
To identify the genes and biological pathways involved in complex immune-related diseases, Ishigaki and his team—led by Kazuhiko Yamamoto and Yuta Kochi, also from the RIKEN Center for Integrative Medical Sciences—first looked for DNA sequences that explained gene activity levels in five kinds of immune cells as well as unfractionated peripheral blood. They used blood samples from 105 healthy Japanese individuals to build their ‘expression quantitative trait loci’ (eQTL) database for each of these cell types.
The researchers then cross-referenced their database against a catalogue of published gene-sleuthing studies and showed that even some non-autoimmune diseases, such as Parkinson’s disease and schizophrenia, have a large immune component. “By investigating critical immune pathways in these diseases, we might be able to develop novel immunologic treatments for them,” Ishigaki says.
 Three of the researchers involved in the present study. From left: Kazuhiko Yamamoto, who heads the Laboratory for Autoimmune Diseases at RIKEN, Kazuyoshi Ishigaki and Yuta Kochi. © 2017 RIKEN Center for Integrative Medical Sciences
Three of the researchers involved in the present study. From left: Kazuhiko Yamamoto, who heads the Laboratory for Autoimmune Diseases at RIKEN, Kazuyoshi Ishigaki and Yuta Kochi. © 2017 RIKEN Center for Integrative Medical Sciences
The RIKEN investigators next applied their cell-sorting model to genetics datasets from thousands of Japanese individuals with rheumatoid arthritis. They found that the combined action of 176 genes altered TNF signaling in a particular subset of immune cells known as helper T cells, which express a surface protein called CD4, but not in other types of immune cells.
According to Ishigaki, drug developers can use this finding to build a new generation of TNF-targeted drugs that target both TNF and CD4 receptors. This will give the drugs a more anti-inflammatory effect as they will act only in those cells that have gone awry in the autoimmune disease. “Our findings suggest that we can improve the efficiency of current anti-TNF therapy,” he says.
Kochi hopes the research community will embrace his team’s eQTL database for identifying genes implicated in particular cells in any disease of interest—and since “the prediction models are provided as supplemental data in the article,” he says, “anyone can use them.”
Related content
References
- 1. Ishigaki, K., Kochi, Y., Suzuki, A., Tsuchida, Y., Tsuchiya, H., Sumitomo, S., Yamaguchi, K., Nagafuchi, Y., Nakachi, S., Kato, R. et al. Polygenic burdens on cell-specific pathways underlie the risk of rheumatoid arthritis. Nature Genetics 49, 1120–1125 (2017). doi: 10.1038/ng.3885
