Jul. 6, 2018 Research Highlight Biology
A hexameric ion-channel complex revs up a bacterial engine
A bacterial engine turns on and off as the number of its protein units changes
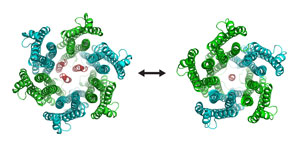 Figure 1: The hexameric (left) and pentameric (right) structures of the ExbB/ExbD complex in the Ton system obtained from x-ray crystallography and single-particle cryo-electron microscopy. ExbB subunits are shown in cyan and green, and ExbD in red. © 2018 RIKEN SPring-8 Center
Figure 1: The hexameric (left) and pentameric (right) structures of the ExbB/ExbD complex in the Ton system obtained from x-ray crystallography and single-particle cryo-electron microscopy. ExbB subunits are shown in cyan and green, and ExbD in red. © 2018 RIKEN SPring-8 Center
The two structures of a protein complex that provides energy for transporting materials across bacterial membranes have been elucidated by RIKEN researchers1. This will help deepen our understanding of how to use a fundamental energy source, the proton motive force, in living organisms.
Many essential biological processes are powered by ions flowing across cell membranes. A protein complex known as the Ton system in the inner membrane of many bacteria harnesses the energy generated when protons flow into the cell to transport compounds, including essential nutrients, across the outer membrane. Determining the complex’s structure and function could help researchers develop drugs that attach to pathogenic bacteria.
The Ton complex is formed from three proteins: ExbB, ExbD and TonB. Researchers have clarified some aspects of its structure and function, but its working mechanisms remain unclear. “The ExbB/ExbD complex is a challenging target as it is difficult to crystallize for x-ray diffraction and relatively small for single-particle cryo-electron microscopy (cryo-EM),” explains Koji Yonekura of the RIKEN SPring-8 Center.
By combining both techniques, Yonekura’s team gained a more detailed understanding of the proteins’ structures. They separated ExbB and ExbD proteins from bacterial membranes and were able to obtain sample crystals that gave good x-ray diffraction images for ExbB but not for ExbD. The researchers also placed the ExbB/ExbD protein complexes in different levels of acidity, froze them and obtained cryo-EM images, from which they generated three-dimensional models of the protein complex for both ExbB and ExbD.
Scientists have long debated how many ExbB and ExbD subunits are in the Ton complex. Yonekura’s team discovered that the numbers vary depending on the local acidity in the bacterial membrane.
In higher acidity, the ExbB/ExbD complex consists of five ExbB (pentamer) and one ExbD subunits. In contrast, in lower acidity, the complex consists of six ExbB (hexamer) and three ExbD subunits (Fig. 1). The hexamer complex represents an open, active channel, whereas the narrower pentamer channel represents an inactive state. This is reasonable since accumulating too many protons inside a cell is toxic to bacteria. This mechanism could be used to activate and inactivate this system.
The team hypothesizes that protons move through the spiral-shaped six ExbB proteins, generating a torsional force that rotates the three ExbD subunits. This energy is transferred to the TonB protein part of the complex, which reaches out and connects to receptors on the outer layer of the bacterial cell membrane. This connection changes the receptors’ structures such that they release substances bound to them for transfer into the cell.
“Our findings could provide a new vision of dynamic membrane biology,” says Yonekura.
Related contents
- Structural insights into a protein-modification mechanism that boosts bacterial virulence
- How bacteria avoid poisoning themselves
- Unraveling how bacteria motor along
References
- 1. Maki-Yonekura, S., Matsuoka, R., Yamashita, Y., Shimizu, H., Tanaka, M., Iwabuki, F. & Yonekura, K. Hexameric and pentameric complexes of the ExbBD energizer in the Ton system. eLife 7, e35419 (2018). doi: 10.7554/eLife.35419
