Sep. 7, 2018 Research Highlight Biology
100 genes linked to inflammatory bowel disease by post-GWAS method
A new technique for analyzing the genetics of diseases has enabled scientists to get closer to finding the genes responsible for inflammatory bowel disease
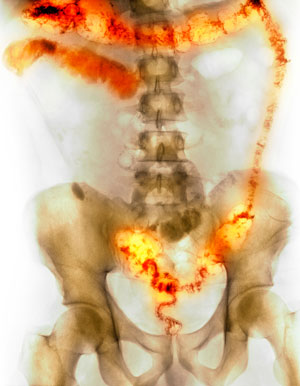 Figure 1: Colored x-ray image showing the inflamed and ulcerated colon of a man with inflammatory bowel disease. Severe cases such as this sometimes necessitate surgical removal of the colon. © SCIENCE PHOTO LIBRARY
Figure 1: Colored x-ray image showing the inflamed and ulcerated colon of a man with inflammatory bowel disease. Severe cases such as this sometimes necessitate surgical removal of the colon. © SCIENCE PHOTO LIBRARY
By conducting a genetic analysis on 350 people, RIKEN geneticists have narrowed down the search for gene variants that give rise to inflammatory bowel disease1. This information will help researchers find ways to combat the disease.
A few diseases, such as cystic fibrosis and sickle cell disease, are caused by a variant of a single gene. But the vast majority of diseases have far more complex genetic causes, with often hundreds or thousands of genes that could give rise to them.
A powerful tool for exploring the genetics of a disease is the genome-wide association study (GWAS). It compares the genomes of people with a certain disease to those of healthy individuals and identifies small variations in DNA that are more prevalent in people with the disease. But a GWAS cannot pinpoint single genes—it only identifies regions of DNA containing about 5 to 50 genes.
To overcome this limitation, Yukihide Momozawa of the RIKEN Center for Integrative Medical Science and his team are developing more-accurate genetic techniques.
Inflammatory bowel disease is a chronic condition that involves inflammation of the intestinal walls and causes symptoms such as bloody diarrhea and abdominal pain. GWASs have linked over 200 regions of DNA to the disease. Most of these regions do not actually code for proteins; instead they turn other genes, known as causative genes, on and off. These causative genes are the real culprits behind inflammatory bowel disease.
 Yukihide Momozawa and colleagues have used a new genetic analysis technique to identify about 100 probable causative genes for inflammatory bowel disease. © 2018 RIKEN
Yukihide Momozawa and colleagues have used a new genetic analysis technique to identify about 100 probable causative genes for inflammatory bowel disease. © 2018 RIKEN
Now, Momozawa and colleagues have used a new genetic analysis technique to investigate the expression of about 2,000 genes located in the 200 regions identified by GWAS. This enabled them to find about 100 probable causative genes that are very likely to be directly involved in triggering inflammatory bowel disease. These genes could be targeted by future drugs, although Momozawa cautions that much work still needs to be done for that to become a reality.
The study also uncovered a new plot twist. “The story turns out to be more complicated than previously thought,” comments Momozawa. “We found that sometimes a single variant influences two or more genes significantly more often in the 200 regions. This was an unexpected and new result.”
Momozawa and his team want to apply the same technique to other diseases. “We want to look at other diseases, such as colon cancer or other autoimmune diseases such as rheumatoid arthritis and Graves’ disease, and to generate a huge database that will help in the development of genetics tests for patients.”
Related contents
- Global genes for gut disorders
- Meta-analysis of genome-wide association studies triples known genetic risk factors for stroke
- A healthy balance
References
- 1. Momozawa, Y, Dmitrieva, J., Théâtre, E., Deffontaine, V., Rahmouni, S., Charloteaux, B., Crins, F., Docampo, E., Elansary, M., Gori, A.-S. et al. IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nature Communications 9, 2427 (2018). doi: 10.1038/s41467-018-04365-8
