Nov. 15, 2019 Research Highlight Biology
Detecting the metabolites of a single cell
By using an ultrasensitive analysis technique, scientists can now detect the small molecules that a single cell makes
 Figure 1: A fluorescence micrograph of HeLa cells—cervical cancer cells belonging to the first human cell line. © DR TORSTEN WITTMANN/SCIENCE PHOTO LIBRARY
Figure 1: A fluorescence micrograph of HeLa cells—cervical cancer cells belonging to the first human cell line. © DR TORSTEN WITTMANN/SCIENCE PHOTO LIBRARY
An analytical technique that is so sensitive it can identify the small molecules produced by a single cell has been made and demonstrated by an all-RIKEN team1. This opens up the possibility of analyzing the roles individual cancer cells play in tumor growth and survival.
Cells are tiny chemical factories, constantly manufacturing the compounds they need to survive and thrive. Their DNA contains instructions for how to make these compounds. Which compounds they produce at any particular time is determined in part by their environment.
Geneticists can currently determine the genetic makeup of a single cell and the messenger RNA that it produces. But there is no technique capable of reliably profiling the small molecules, or metabolites, that a single cell produces. Researchers would dearly love to be able to do this because it would provide a valuable window into how single cells react to their environment.
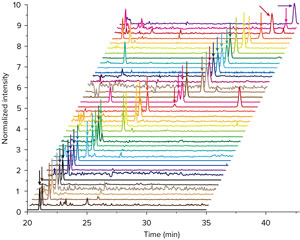 Figure 2: A RIKEN team demonstrated the sensitivity of their metabolomic analysis technique by using it to detect 450 metabolites (40 identified; to see their names, click on the image to bring up a larger, labeled image) from a single HeLa cell. Reprinted, with permission, from Ref. 1. Copyright (2019) American Chemical Society
Figure 2: A RIKEN team demonstrated the sensitivity of their metabolomic analysis technique by using it to detect 450 metabolites (40 identified; to see their names, click on the image to bring up a larger, labeled image) from a single HeLa cell. Reprinted, with permission, from Ref. 1. Copyright (2019) American Chemical Society
One area where such a technique is badly needed is cancer biology. “Cancer stem cells regulate their environment so that it is acidic and has low oxygen levels—this is a vital survival strategy for cancer cells,” explains Takayuki Kawai of the RIKEN Center for Biosystems Dynamics Research. “But since this is a microscale phenomenon involving 10 to 100 cells, it is important to analyze microscale samples, even down to a single-cell level.”
Now, Kawai and his co-workers have boosted the sensitivity of a conventional metabolomic analytic technique known as capillary electrophoresis–mass spectrometry (CE-MS) by up to 800 times, allowing them to profile the metabolites that a single cell produces.
The researchers improved CE-MS, which is typically used to analyze tens of thousands of cells, in two ways—they used a specially designed system to concentrate the sample before analysis and they used a thin-walled, tapered glass capillary tube to deliver the sample to the mass spectrometer.
 Takayuki Kawai and his co-workers have developed an ultrasensitive metabolomic analysis technique based on capillary electrophoresis−mass spectrometry that can detect the metabolites of a single cell. © 2019 RIKEN
Takayuki Kawai and his co-workers have developed an ultrasensitive metabolomic analysis technique based on capillary electrophoresis−mass spectrometry that can detect the metabolites of a single cell. © 2019 RIKEN
The team demonstrated the sensitivity of their technique by using it to identify 40 metabolites from a single human cervical cancer cell (Fig. 2). “This is the first time this level of analysis has been achieved,” notes Kawai. “That’s because our system is currently the most sensitive in the metabolomic analysis system in the world.”
The team is looking for ways to apply their technique to areas of clinical interest. “This technique allows us to directly analyze metabolites from cellular microenvironments in humans such as tumors,” says Kawai. “We’re now searching for good target molecules for clinical applications, such as drug discovery and diagnosis.”
Related contents
- What’s in this plant? The best automated system for finding potential drugs
- Getting a picture of the molecules in a cell in just minutes
Reference
- 1. Kawai, T., Ota, N., Okada, K., Imasato, A., Owa, Y., Morita, M., Tada, M. & Tanaka, Y. Ultrasensitive single cell metabolomics by capillary electrophoresis−mass spectrometry with a thin-walled tapered emitter and large-volume dual sample preconcentration. Analytical Chemistry 91, 10564–10572 (2019). doi: 10.1021/acs.analchem.9b01578
