Sep. 25, 2020 Perspectives Biology
Japan’s big brain project: advances light up marmoset brains
RIKEN researchers are coordinating Japan’s Brain/MINDS project and illuminating marmoset neural networks—adding key primate brain data to other large-scale brain-mapping efforts.
 Image of a marmoset © BlackCatPhotos / Getty Images
Image of a marmoset © BlackCatPhotos / Getty Images
Our knowledge of brain neural connections remains remarkably incomplete. To date, the nematode worm Caenorhabditis elegans is the only species that boasts a complete wiring diagram of its neurons.
While the last decade of studies have revealed insights into neurological and psychiatric diseases (such as Alzheimer’s and Parkinson’s diseases and schizophrenia), existing data is often scattered and incomplete. As a result, many neuroscience groups are now creating three-dimensional digital brain atlases that collate knowledge on mechanisms of brain activity to inform future experimental research.
In Japan, one of the largest projects of this kind is the Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) project, coordinated by the RIKEN Center for Brain Science (CBS). Launched in 2014, the 10 year, 40 billion yen (US$350 million), multi-institutional project is developing mapping technologies and digitizing brain data, with a focus on primates.
Japan’s effort is a parallel to the large-scale brain mapping efforts launched in 2013 through the European Union’s €1-billion (US$1.3-billion) Human Brain Project (HPB) and the United States’ US$1-billion Brain Research through Advancing Innovative Neurotechnologies Initiative (BRAIN). Each of these efforts has taken a slightly different approach: the HBP started as centralized enterprise with a computational focus, aimed at building detailed models of neural circuitry, whereas the BRAIN Initiative initially had more of an emphasis on the development of technologies to facilitate neuroscience research. An equivalent China Brain Project was launched in 2019.
While Japan’s effort draws on a smaller funding pool than some of these huge initiative, Brain/MINDs has access to an in-demand and globally scarce resource—a population of teacup-sized, genetically modified marmosets (Callithrix jacchus, left) that have been developed over more than ten years by Erika Sasaki and her colleagues at the Central Institute for Experimental Animals in Kawasaki in collaboration with Hideyuki Okano at the Keio University School of Medicine.
The USA’s research populations of these primates has tripled over the last ten years due to the fact that marmosets give birth more often and age faster than other common primate research models. This speeds up studies, particularly those on aging brains. In addition, the marmoset cortex structure is less folded than many other primate models, making it easier to image neuronal networks.
Marmosets also share many human developmental processes and anatomical structures. They boast similar social behaviors, including strong relationships between parents and offspring, a social vocal communication method, as well as parallel neurological diseases. As a result, data from marmosets fill a crucial research gap between disease models in common smaller experimental animals, such as mice, which often fail to mimic human brain disorders, and models of the human brain that need extensive and expensive data validation.
Lighting up the marmoset connectome
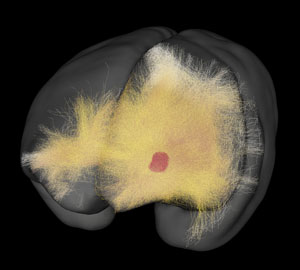 A 3D map of neural networks created by Tetsuo Yamamori’s team can be explored on the Brain/MINDS website: Brain/MINDS Data Portal © 2020 RIKEN
A 3D map of neural networks created by Tetsuo Yamamori’s team can be explored on the Brain/MINDS website: Brain/MINDS Data Portal © 2020 RIKEN
Currently, the Brain/MINDS project (which is co-led by RIKEN’s Hideyuki Okano and Atsushi Miyawaki) is linking detailed data on brain physiology with macro-scale functional analysis. The hope is to improve the diagnosis and treatment of psychiatric and neurological disorders by drawing links between chemical and neural network activation and function. The project is also continuing to map brains, establish new mapping technologies and conduct clinical research.
At CBS, the Molecular Analysis of Higher Brain Function team is performing intermediate-scale mapping of the marmoset prefrontal cortex, a part of the neocortex, the outer layer of the brain unique to mammals (see "Meet the brain region that may control your consciousness" for another RIKEN breakthrough on the neocortex). The neocortex is linked to high-level cognition, personality expression, decision making and the moderation of social behaviour.
Intermediate-scale brain mapping employs chemical tracers and light microscopy to show the structure of neurons and neural networks, revealing brain-wide connectivity and inter-area neuron activity. This type of mapping falls between macroscale mapping of brain region activation performed largely by MRIs and microscale mapping of tissue changes using electron-microscopic analysis.
The technique used by the team mirrors the tracer technology employed by the Allen Mouse Brain Connectivity Atlas, launched in 2014. However, the Molecular Analysis of Higher Brain Function team has developed a way to enhance tracer signals that is better able to map marmoset brains in live animals over the long-term compared to tracers developed for smaller model animals, such as mice, fish and flies.
A fluorescence protein gene is carried by an adeno-associated virus vector that is injected into brain tissue. These viruses also carry genes that enhance the detection of calcium in neurons by increasing a protein called "GCaMP", which activates the fluorescent proteins, lighting up neurons to their axial ends and across synapses. A TissueCyte machine can then perform a layered imaging technique known as serial two-photon (STP) tomography to help build a 3D image of axonal spread from the injection site.
In 2015, the group published on the successful use of this tet-tre GCaMP fluorescence tracer system in the primate cortex for the first time. GCaMP highlights the calcium, a charge carrier and an intracellular messenger in neurons that’s important for neuron growth, and synaptic and cognitive function. In the paper, the group detailed the use of the tet-tre GCaMP system to help achieve long-term two-photon neuronal imaging of marmoset brains, which was able to be detected for more than 100 days after injection, and showed responses to tactile stimulation imaged at subcellular resolution.
This method has since been widely used by other Brain/MINDS teams, including the Matsuzaki Lab, Ohki Lab and Okano Lab. In 2017, Keio University and CBS’s Laboratory for Marmoset Neural Architecture (Okano Lab) used it to image neurons linked to marmosets moving levers with their arms and ladder climbing. In 2018, CBS’s Brain Functional Dynamics Collaboration Laboratory (Matsuzaki Lab) used the technology to show the neurons activated by marmosets moving cursors on screens. It has also been used in a number of key experiments in laboratories beyond Brain/MINDs.
A separate imaging system has been developed by Brain/MINDS in CBS’s Atsushi Miyawaki Lab. In 2018, the group reported a bioluminescent protein that strongly responds to long (red) wavelengths of light, genetically engineered from firefly luciferase (the light-emitting enzyme responsible for bioluminescence in fireflies and click beetles). This protein is able to detect deep-tissue signals at a single-cell level in freely moving animals and create a brighter emission by up to a factor of 1,000 compared to conventional technology.
Using this technique, the Molecular Analysis of Higher Brain Function team and the Miyawaki Lab were able to detect and image small numbers of striatal neurons in the brains of naturally behaving marmosets.
Digital brain simulations
A parallel challenge for Brain/MINDS is dealing with detailed 3D images of brain connections that can be hundreds of gigabytes in size. To tackle this, the Brain Image Analysis Unit is developing algorithms that process and automatically annotate marmoset brain images, which will allow efficient integration of a huge amount of imaging data from different Brain/MINDS research groups.
In addition, the Connectome Analysis Unit is making this data accessible through the Brain/MINDS online data portal, and it is hoped that interactive web pages for the prefrontal cortex tracer studies mentioned previously will be made public within the coming year. These pages will allow the exploration of very high-resolution image data to easily see real connectivity patterns between brain regions. For example, researchers can map an atlas to new experimental data and carry out brain region to brain region analysis.
This will complement previous atlases developed by Brain/MINDS, including a 3D digital atlas of the marmoset brain based on macro-scale MRI data and micro-scale histology imaging (using staining of Nissl bodies, small granular bodies in neurons). Created in 2018, in collaboration with neuroanatomist Tsutomu Hashikawa, this atlas can be integrated with popular neuroscience tools. The new tracer map will help connect macro-scale functional insights and micro-scale tissue changes to neural networks.
Digital advances will be key to addressing criticisms of existing brain mapping projects that note that brain simulations aren’t being finalized as fast as predicted. Nonetheless, some projects are reporting important real-world results. For example, in 2019 the UK’s HBP announced clinical trials for a brain simulator that can individualize therapeutic surgery for epilepsy patients.
The human brain has more than 15 billion neurons, so creating insights that have clinical applications is a mammoth task. However, this only underlines the case for marmoset brain maps, which should help narrow the search for relevant neural networks in humans. We anticipate that our digitized brain atlases will be important resources to the neuroscience community.
References
About the Researchers
Tetsuo Yamamori, Team Leader, Molecular Analysis of Higher Brain Function, RIKEN Center for Brain Science
Tetsuo Yamamori joined RIKEN in 2014 as a team leader for the RIKEN Brain Science Institute. Prior to this he was Deputy Director-General of National Institute for Basic Biology in Japan. He had previously worked as a research fellow in the RIKEN Frontier Research Program (1991 to 1994). He is an associate editor for Frontiers in Neuroanatomy.
Henrik Skibbe, Unit Leader, Brain Image Analysis Unit, RIKEN Center for Brain Science

Henrik Skibbe joined RIKEN in 2019. Previously he had been a Specially Appointed Assistant Professor at Kyoto University. Skibbe moved to Japan from Germany in 2013 to work as a postdoctoral researcher at Kyoto University. In Germany, he worked in medical physics at the University Hospital Freiburg and in advanced computing at the University of Freiburg.
Alexander Woodward, Unit Leader, Analysis Unit, RIKEN Center for Brain Science

Alexander Woodward joined RIKEN in 2015. Between 2010 and 2015 he was a Japan Society for the Promotion of Science (JSPS) Post Doctoral Fellow at the University of Tokyo. He gained a PhD in computer vision, a field of AI that trains computers to interpret and understand the visual world, at the University of Auckland in New Zealand.
