Oct. 21, 2021 Research Highlight Medicine / Disease
Supercomputer simulations reveal how protein crowding in cells impacts interactions
Microsecond-scale simulations are helping scientists understand how drugs ‘navigate the crowd’ inside cells
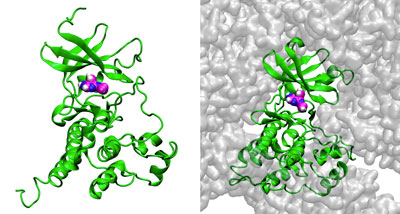 Figure 1: Simulations of the interaction of the enzyme c-Src kinase (green) with inhibitor PP1 (magenta, blue and white spheres) in the presence of low (left) and high (right) concentrations of bovine serum albumin (BSA; gray). The simulations reveal that crowding by BSA reduces the ability of PP1 to bind with c-Src kinase. © LAGUNA DESIGN/SCIENCE PHOTO LIBRARY
Figure 1: Simulations of the interaction of the enzyme c-Src kinase (green) with inhibitor PP1 (magenta, blue and white spheres) in the presence of low (left) and high (right) concentrations of bovine serum albumin (BSA; gray). The simulations reveal that crowding by BSA reduces the ability of PP1 to bind with c-Src kinase. © LAGUNA DESIGN/SCIENCE PHOTO LIBRARY
Supercomputer simulations by RIKEN researchers have revealed how drug binding to a protein target changes as the surrounding environment becomes more cluttered with other proteins1. These simulations could help improve drug development since they shed light on why some drugs work in theory but flop in practice.
The initial stages of drug development usually involve simulating the interactions between a molecule and its target protein. Although these simulations can suggest a drug is effective, the interaction can frequently fail to live up to its promise when tested on living cells. Computational biophysicist Yuji Sugita of the RIKEN Center for Biosystems Dynamics Research (BDR) and his colleagues wanted to make simulations more accurate by accounting for cells’ normally crowded environments.
“Many proteins and macromolecules are present inside cells, making it a crowded environment,” explains Sugita. “In fact, large molecules account for anywhere between a fourth and a half of the volume of a cell’s cytoplasm. We wanted to discover how these crowded environments affect drug binding to target proteins.”
To do this, they developed a highly optimized software program called GENESIS for use with supercomputers in Japan. They then conducted microsecond-scale simulations of the interaction of an enzyme (c-Src kinase) with an inhibitor (PP1) in the presence of different concentrations of bovine serum albumin (BSA). Sugita’s collaborator, Michael Feig, a professor at Michigan State University, also performed simulations of the same systems using a molecular-dynamics supercomputer in the United States. The team chose c-Src kinase because the enzyme regulates signal transduction pathways and its dysregulation is associated with many diseases, including cancers.
The researchers found that crowding by BSA reduced the amount of PP1 able to reach the enzyme by physically blocking its access and also by weakly and non-specifically interacting with it.
Still, small amounts of PP1 were able to slip through the crowds. But the simulations showed that BSA crowding also covered some binding sites on c-Src kinase and changed the enzyme’s shape, altering the pathways available for it to reach its main binding site.
The team validated their results by performing laboratory tests using the actual proteins in similar conditions. These experiments conducted by Mikako Shirouzu and her colleagues in RIKEN BDR showed that PP1’s efficacy in inhibiting c-Src kinase decreased with increasing BSA crowding.
The team now wants to examine how crowding affects other target proteins and drugs. They also intend to use their supercomputers to study protein function inside biological membranes and cell organelles. “These different environments could affect protein functions similarly or differently,” says Sugita. “We just don’t know.”
Related contents
- Basement membrane underpins tissue interactions in the skin
- A nuclear ‘gatekeeper’ that regulates bone formation
- Precision-medicine approach turns up novel drug combination for aggressive leukemias
Reference
- 1. Kasahara, K., Re, S., Nawrocki, G., Oshima, H., Mishima-Tsumagari, C. Miyata-Yabuki, Y., Kukimoto-Niino, M., Yu, I., Shirouzu, M., Feig, M. et al. Reduced efficacy of a Src kinase inhibitor in crowded protein solution. Nature Communications 12, 4099 (2021). doi: 10.1038/s41467-021-24349-5
